Natural Product Target Network Reveals Potential for Cancer Combination Therapies
- PMID: 31214023
- PMCID: PMC6555193
- DOI: 10.3389/fphar.2019.00557
Natural Product Target Network Reveals Potential for Cancer Combination Therapies
Abstract
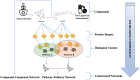
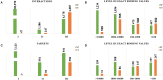
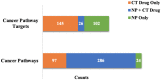
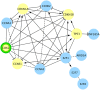
A body of research demonstrates examples of in vitro and in vivo synergy between natural products and anti-neoplastic drugs for some cancers. However, the underlying biological mechanisms are still elusive. To better understand biological entities targeted by natural products and therefore provide rational evidence for future novel combination therapies for cancer treatment, we assess the targetable space of natural products using public domain compound-target information. When considering pathways from the Reactome database targeted by natural products, we found an increase in coverage of 61% (725 pathways), relative to pathways covered by FDA approved cancer drugs collected in the Cancer Targetome, a resource for evidence-based drug-target interactions. Not only is the coverage of pathways targeted by compounds increased when we include natural products, but coverage of targets within those pathways is also increased. Furthermore, we examined the distribution of cancer driver genes across pathways to assess relevance of natural products to critical cancer therapeutic space. We found 24 pathways enriched for cancer drivers that had no available cancer drug interactions at a potentially clinically relevant binding affinity threshold of < 100nM that had at least one natural product interaction at that same binding threshold. Assessment of network context highlighted the fact that natural products show target family groupings both distinct from and in common with cancer drugs, strengthening the complementary potential for natural products in the cancer therapeutic space. In conclusion, our study provides a foundation for developing novel cancer treatment with the combination of drugs and natural products.
Keywords: antineoplastic drug; cancer; natural product; synergy; therapeutic targets.
Figures
References
-
- Becker M. S., Schmezer P., Breuer R., Haas S. F., Essers M. A., Krammer P. H., et al. . (2014). The traditional Chinese medical compound Rocaglamide protects nonmalignant primary cells from DNA damage-induced toxicity by inhibition of p53 expression. Cell Death Dis. 5:e1000. 10.1038/cddis.2013.528 - DOI - PMC - PubMed
-
- Benjamini Y., Yekutieli D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188. 10.1214/aos/1013699998 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

