Simultaneous Colorimetric Detection of a Variety of Salmonella spp. in Food and Environmental Samples by Optical Biosensing Using Oligonucleotide-Gold Nanoparticles
- PMID: 31214132
- PMCID: PMC6554661
- DOI: 10.3389/fmicb.2019.01138
Simultaneous Colorimetric Detection of a Variety of Salmonella spp. in Food and Environmental Samples by Optical Biosensing Using Oligonucleotide-Gold Nanoparticles
Abstract
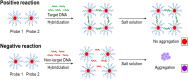
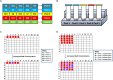
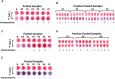
Optical biosensors for rapid detection of significant foodborne pathogens are steadily gaining popularity due to its simplicity and sensitivity. While nanomaterials such as gold nanoparticles (AuNPs) are commonly used as signal amplifiers for optical biosensors, AuNPs can also be utilized as a robust biosensing platform. Many reported optical biosensors were designed for individual pathogen detection in a single assay and have high detection limit (DL). Salmonella spp. is one of the major causative agents of foodborne sickness, hospitalization and deaths. Unfortunately, there are around 2,000 serotypes of Salmonella worldwide, and rapid and simultaneous detection of multiple strains in a single assay is lacking. In this study, a comprehensive and highly sensitive simultaneous colorimetric detection of nineteen (19) environmental and outbreak Salmonella spp. strains was achieved by a novel optical biosensing platform using oligonucleotide-functionalized AuNPs. A pair of newly designed single stranded oligonucleotides (30-mer) was displayed onto the surface of AuNPs (13 nm) as detection probes to hybridize with a conserved genomic region (192-bases) of ttrRSBCA found on a broad range of Salmonella spp. strains. The sandwich hybridization (30 min, 55°C) resulted in a structural formation of highly stable oligonucleotide/AuNPs-DNA complexes which remained undisturbed even after subjecting to an increased salt concentration (2 M, final), thus allowing a direct discrimination via color change of target (red color) from non-target (purplish-blue color) reaction mixtures by direct observation using the naked eye. In food matrices (blueberries and chicken meat), nineteen different Salmonella spp. strains were concentrated using immunomagnetic separation and then simultaneously detected in a 96-well microplate by oligonucleotide-functionalized AuNPs after DNA preparation. Successful oligonucleotide/AuNPs-DNA hybridization was confirmed by gel electrophoresis while AuNPs aggregation in non-target and control reaction mixtures was verified by both spectrophotometric analysis and TEM images. Results showed that the optical AuNP biosensing platform can simultaneously screen nineteen (19) viable Salmonella spp. strains tested with 100% specificity and a superior detection limit of <10 CFU/mL or g for both pure culture and complex matrices setups. The highly sensitive colorimetric detection system can significantly improve the screening and detection of viable Salmonella spp. strains present in complex food and environmental matrices, therefore reducing the risks of contamination and incidence of foodborne diseases.
Keywords: Salmonella; colorimetric; gold nanoparticles; oligonucleotides; optical biosensor; ttrRSBCA.
Figures


References
-
- Abdelhaseib M. U., Singh A. K., Bailey M., Singh M., El-Khateib T., Bhunia A. K. (2016). Fiber optic and light scattering sensors: complimentary approaches to rapid detection of Salmonella enterica in food samples. Food Control 61 135–145.
-
- Ahn S., Walt D. R. (2005). Detection of Salmonella spp. using microsphere-based, fiber-optic DNA microarrays. Anal. Chem. 77 5041–5047. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

