Integrating Clinical and Genetic Analysis of Perineural Invasion in Head and Neck Squamous Cell Carcinoma
- PMID: 31214495
- PMCID: PMC6555133
- DOI: 10.3389/fonc.2019.00434
Integrating Clinical and Genetic Analysis of Perineural Invasion in Head and Neck Squamous Cell Carcinoma
Abstract
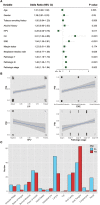
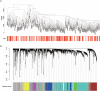
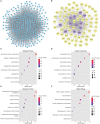
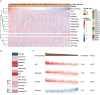
Introduction: Perineural invasion (PNI), a key pathological feature of head and neck squamous cell carcinoma (HNSCC), predicts poor survival. However, the associated clinical characteristics remain uncertain, and the molecular mechanisms are largely unknown. Materials and methods: HNSCC gene expression and corresponding clinical data were downloaded from The Cancer Genome Atlas (TCGA). Prognostic subgroup analysis was performed, and potential PNI risk factors were assessed with logistic regression. PNI-associated gene coexpression modules were identified with weighted gene coexpression network analysis (WGCNA), and key module gene functions and the roles of non-malignant cells in PNI were evaluated with a single-cell transcriptomic dataset (GSE103322). Results: PNI was significantly inversely associated with overall survival (HR, 2.08; 95% CI, 1.27 to 3.40; P = 0.004), especially in advanced patients (HR, 2.62; 95% CI, 1.48 to 4.64; P < 0.001). Age, gender, smoking history, and alcohol history were not risk factors. HPV-positive cases were less likely than HPV-negative cases to develop PNI (OR, 0.28; 95% CI, 0.09 to 0.76; P = 0.017). WGCNA identified a unique significantly PNI-associated coexpression module containing 357 genes, with 12 hub genes (TIMP2, MIR198, LAMA4, FAM198B, MIR4649, COL5A1, COL1A2, OLFML2B, MMP2, FBN1, ADAM12, and PDGFRB). Single-cell transcriptomic data analysis revealed that the genes in the PNI-associated module correlated with the signatures "EMT," "metastasis," and "invasion." Among non-malignant cells, fibroblasts had relatively high expression of the key genes. Conclusion: At the molecular and omic levels, we verified that PNI in HNSCC is a process of invasion rather than simple diffusion. Fibroblasts probably play an important role in PNI. Novelty & Impact Statements The study is a thorough analysis of PNI in HNSCC from the clinical level to the molecular level and presents the first description of cancer-related PNI from the omics perspective to date as far as we know. We verified that PNI in HNSCC is a process of invasion rather than simple diffusion, at the molecular and omic levels. Fibroblasts were found to probably play an important role in PNI by analyzing single-cell transcriptomic data.
Keywords: HNSCC; TCGA; WGCNA; perineural invasion; single-cell sequencing.
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous

