Prediction of the Antioxidant Response Elements' Response of Compound by Deep Learning
- PMID: 31214568
- PMCID: PMC6554289
- DOI: 10.3389/fchem.2019.00385
Prediction of the Antioxidant Response Elements' Response of Compound by Deep Learning
Abstract
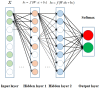
The antioxidant response elements (AREs) play a significant role in occurrence of oxidative stress and may cause multitudinous toxicity effects in the pathogenesis of a variety of diseases. Determining if one compound can activate AREs is crucial for the assessment of potential risk of compound. Here, a series of predictive models by applying multiple deep learning algorithms including deep neural networks (DNN), convolution neural networks (CNN), recurrent neural networks (RNN), and highway networks (HN) were constructed and validated based on Tox21 challenge dataset and applied to predict whether the compounds are the activators or inactivators of AREs. The built models were evaluated by various of statistical parameters, such as sensitivity, specificity, accuracy, Matthews correlation coefficient (MCC) and receiver operating characteristic (ROC) curve. The DNN prediction model based on fingerprint features has best prediction ability, with accuracy of 0.992, 0.914, and 0.917 for the training set, test set, and validation set, respectively. Consequently, these robust models can be adopted to predict the ARE response of molecules fast and accurately, which is of great significance for the evaluation of safety of compounds in the process of drug discovery and development.
Keywords: antioxidant response elements (AREs); deep learning; machine learning; prediction; toxicity.
Figures






Similar articles
-
Assessment of Automated Identification of Phases in Videos of Cataract Surgery Using Machine Learning and Deep Learning Techniques.JAMA Netw Open. 2019 Apr 5;2(4):e191860. doi: 10.1001/jamanetworkopen.2019.1860. JAMA Netw Open. 2019. PMID: 30951163 Free PMC article.
-
Deep neural networks for human microRNA precursor detection.BMC Bioinformatics. 2020 Jan 13;21(1):17. doi: 10.1186/s12859-020-3339-7. BMC Bioinformatics. 2020. PMID: 31931701 Free PMC article.
-
Deep Learning on High-Throughput Transcriptomics to Predict Drug-Induced Liver Injury.Front Bioeng Biotechnol. 2020 Nov 27;8:562677. doi: 10.3389/fbioe.2020.562677. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 33330410 Free PMC article.
-
Development and Validation of a Deep Learning Radiomics Model Predicting Lymph Node Status in Operable Cervical Cancer.Front Oncol. 2020 Apr 15;10:464. doi: 10.3389/fonc.2020.00464. eCollection 2020. Front Oncol. 2020. PMID: 32373511 Free PMC article.
-
Deep learning in prediction of intrinsic disorder in proteins.Comput Struct Biotechnol J. 2022 Mar 8;20:1286-1294. doi: 10.1016/j.csbj.2022.03.003. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35356546 Free PMC article. Review.
Cited by
-
DeepSnap-Deep Learning Approach Predicts Progesterone Receptor Antagonist Activity With High Performance.Front Bioeng Biotechnol. 2020 Jan 22;7:485. doi: 10.3389/fbioe.2019.00485. eCollection 2019. Front Bioeng Biotechnol. 2020. PMID: 32039185 Free PMC article.
-
Sleep and Oxidative Stress: Current Perspectives on the Role of NRF2.Cell Mol Neurobiol. 2024 Jun 25;44(1):52. doi: 10.1007/s10571-024-01487-0. Cell Mol Neurobiol. 2024. PMID: 38916679 Free PMC article. Review.
-
Identification of Drug-Disease Associations Using Information of Molecular Structures and Clinical Symptoms via Deep Convolutional Neural Network.Front Chem. 2020 Jan 10;7:924. doi: 10.3389/fchem.2019.00924. eCollection 2019. Front Chem. 2020. PMID: 31998700 Free PMC article.
-
In Silico Approaches In Carcinogenicity Hazard Assessment: Current Status and Future Needs.Comput Toxicol. 2021 Nov;20:100191. doi: 10.1016/j.comtox.2021.100191. Epub 2021 Sep 23. Comput Toxicol. 2021. PMID: 35368437 Free PMC article.
References
-
- Abdelaziz A., Spahn-Langguth H., Karl-Werner S., Tetko I. V. (2016). Consensus modeling for HTS assays using in silico descriptors calculates the best balanced accuracy in Tox21 challenge. Front. Environ. Sci. 4, 1–12. 10.3389/fenvs.2016.00002 - DOI
-
- Breiman L. (2001). Random forests. Mach. Learn. 45, 5–32. 10.1023/A:1010933404324 - DOI
-
- Dahl G. E., Jaitly N., Salakhutdinov R. (2014). Multi-task neural networks for QSAR predictions. arXiv: 1406.1231. Available online at: https://arxiv.org/pdf/1406.1231.pdf
LinkOut - more resources
Full Text Sources

