Identification of positively selected genes in human pathogenic treponemes: Syphilis-, yaws-, and bejel-causing strains differ in sets of genes showing adaptive evolution
- PMID: 31216284
- PMCID: PMC6602244
- DOI: 10.1371/journal.pntd.0007463
Identification of positively selected genes in human pathogenic treponemes: Syphilis-, yaws-, and bejel-causing strains differ in sets of genes showing adaptive evolution
Abstract
Background: Pathogenic treponemes related to Treponema pallidum are both human (causing syphilis, yaws, bejel) and animal pathogens (infections of primates, venereal spirochetosis in rabbits). A set of 11 treponemal genome sequences including those of five Treponema pallidum ssp. pallidum (TPA) strains (Nichols, DAL-1, Mexico A, SS14, Chicago), four T. p. ssp. pertenue (TPE) strains (CDC-2, Gauthier, Samoa D, Fribourg-Blanc), one T. p. ssp. endemicum (TEN) strain (Bosnia A) and one strain (Cuniculi A) of Treponema paraluisleporidarum ecovar Cuniculus (TPeC) were tested for the presence of positively selected genes.
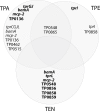
Methodology/principal findings: A total of 1068 orthologous genes annotated in all 11 genomes were tested for the presence of positively selected genes using both site and branch-site models with CODEML (PAML package). Subsequent analyses with sequences obtained from 62 treponemal draft genomes were used for the identification of positively selected amino acid positions. Synthetic biotinylated peptides were designed to cover positively selected protein regions and these peptides were tested for reactivity with the patient's syphilis sera. Altogether, 22 positively selected genes were identified in the TP genomes and TPA sets of positively selected genes differed from TPE genes. While genetic variability among TPA strains was predominantly present in a number of genetic loci, genetic variability within TPE and TEN strains was distributed more equally along the chromosome. Several syphilitic sera were shown to react with some peptides derived from the protein sequences evolving under positive selection.
Conclusions/significance: The syphilis-, yaws-, and bejel-causing strains differed relative to sets of positively selected genes. Most of the positively selected chromosomal loci were identified among the TPA treponemes. The local accumulation of genetic variability suggests that the diversification of TPA strains took place predominantly in a limited number of genomic regions compared to the more dispersed genetic diversity differentiating TPE and TEN strains. The identification of positively selected sites in tpr genes and genes encoding outer membrane proteins suggests their role during infection of human and animal hosts. The driving force for adaptive evolution at these loci thus appears to be the host immune response as supported by observed reactivity of syphilitic sera with some peptides derived from protein sequences showing adaptive evolution.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Whole genome sequences of three Treponema pallidum ssp. pertenue strains: yaws and syphilis treponemes differ in less than 0.2% of the genome sequence.PLoS Negl Trop Dis. 2012 Jan;6(1):e1471. doi: 10.1371/journal.pntd.0001471. Epub 2012 Jan 24. PLoS Negl Trop Dis. 2012. PMID: 22292095 Free PMC article.
-
A Retrospective Study on Genetic Heterogeneity within Treponema Strains: Subpopulations Are Genetically Distinct in a Limited Number of Positions.PLoS Negl Trop Dis. 2015 Oct 5;9(10):e0004110. doi: 10.1371/journal.pntd.0004110. eCollection 2015. PLoS Negl Trop Dis. 2015. PMID: 26436423 Free PMC article.
-
Syphilis-causing strains belong to separate SS14-like or Nichols-like groups as defined by multilocus analysis of 19 Treponema pallidum strains.Int J Med Microbiol. 2014 Jul;304(5-6):645-53. doi: 10.1016/j.ijmm.2014.04.007. Epub 2014 Apr 26. Int J Med Microbiol. 2014. PMID: 24841252
-
Genetic diversity in Treponema pallidum: implications for pathogenesis, evolution and molecular diagnostics of syphilis and yaws.Infect Genet Evol. 2012 Mar;12(2):191-202. doi: 10.1016/j.meegid.2011.12.001. Epub 2011 Dec 15. Infect Genet Evol. 2012. PMID: 22198325 Free PMC article. Review.
-
Genetics of human and animal uncultivable treponemal pathogens.Infect Genet Evol. 2018 Jul;61:92-107. doi: 10.1016/j.meegid.2018.03.015. Epub 2018 Mar 22. Infect Genet Evol. 2018. PMID: 29578082 Review.
Cited by
-
The pan-genome of Treponema pallidum reveals differences in genome plasticity between subspecies related to venereal and non-venereal syphilis.BMC Genomics. 2020 Jan 10;21(1):33. doi: 10.1186/s12864-019-6430-6. BMC Genomics. 2020. PMID: 31924165 Free PMC article.
-
Whole-genome sequencing reveals evidence for inter-species transmission of the yaws bacterium among nonhuman primates in Tanzania.PLoS Negl Trop Dis. 2025 Feb 26;19(2):e0012887. doi: 10.1371/journal.pntd.0012887. eCollection 2025 Feb. PLoS Negl Trop Dis. 2025. PMID: 40009604 Free PMC article.
-
Low genetic diversity of Treponema pallidum ssp. pertenue (TPE) isolated from patients' ulcers in Namatanai District of Papua New Guinea: Local human population is infected by three TPE genotypes.PLoS Negl Trop Dis. 2024 Jan 2;18(1):e0011831. doi: 10.1371/journal.pntd.0011831. eCollection 2024 Jan. PLoS Negl Trop Dis. 2024. PMID: 38166151 Free PMC article.
-
Comparative genomics and full-length Tprk profiling of Treponema pallidum subsp. pallidum reinfection.PLoS Negl Trop Dis. 2020 Apr 6;14(4):e0007921. doi: 10.1371/journal.pntd.0007921. eCollection 2020 Apr. PLoS Negl Trop Dis. 2020. PMID: 32251462 Free PMC article.
-
The hare syphilis agent is related to, but distinct from, the treponeme causing rabbit syphilis.PLoS One. 2024 Aug 12;19(8):e0307196. doi: 10.1371/journal.pone.0307196. eCollection 2024. PLoS One. 2024. PMID: 39133700 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

