Genome-Wide Analysis of the MADS-Box Transcription Factor Family in Solanum lycopersicum
- PMID: 31216621
- PMCID: PMC6627509
- DOI: 10.3390/ijms20122961
Genome-Wide Analysis of the MADS-Box Transcription Factor Family in Solanum lycopersicum
Abstract
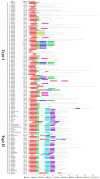
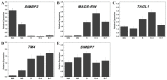
MADS-box family genes encode transcription factors that are involved in multiple developmental processes in plants, especially in floral organ specification, fruit development, and ripening. However, a comprehensive analysis of tomato MADS-box family genes, which is an important model plant to study flower fruit development and ripening, remains obscure. To gain insight into the MADS-box genes in tomato, 131 tomato MADS-box genes were identified. These genes could be divided into five groups (Mα, Mβ, Mγ, Mδ, and MIKC) and were found to be located on all 12 chromosomes. We further analyzed the phylogenetic relationships among Arabidopsis and tomato, as well as the protein motif structure and exon-intron organization, to better understand the tomato MADS-box gene family. Additionally, owing to the role of MADS-box genes in floral organ identification and fruit development, the constitutive expression patterns of MADS-box genes at different stages in tomato development were identified. We analyzed 15 tomato MADS-box genes involved in floral organ identification and five tomato MADS-box genes related to fruit development by qRT-PCR. Collectively, our study provides a comprehensive and systematic analysis of the tomato MADS-box genes and would be valuable for the further functional characterization of some important members of the MADS-box gene family.
Keywords: MADS-box; floral organ; fruit development; genome-wide analysis; tomato.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Genome-wide identification, characterisation and expression analysis of the MADS-box gene family in Prunus mume.Mol Genet Genomics. 2014 Oct;289(5):903-20. doi: 10.1007/s00438-014-0863-z. Epub 2014 May 25. Mol Genet Genomics. 2014. PMID: 24859011
-
MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress.BMC Genomics. 2007 Jul 18;8:242. doi: 10.1186/1471-2164-8-242. BMC Genomics. 2007. PMID: 17640358 Free PMC article.
-
Genome-wide survey of potato MADS-box genes reveals that StMADS1 and StMADS13 are putative downstream targets of tuberigen StSP6A.BMC Genomics. 2018 Oct 3;19(1):726. doi: 10.1186/s12864-018-5113-z. BMC Genomics. 2018. PMID: 30285611 Free PMC article.
-
Variations on a theme in fruit development: the PLE lineage of MADS-box genes in tomato (TAGL1) and other species.Planta. 2017 Aug;246(2):313-321. doi: 10.1007/s00425-017-2725-5. Epub 2017 Jun 28. Planta. 2017. PMID: 28660293 Review.
-
How Hormones and MADS-Box Transcription Factors Are Involved in Controlling Fruit Set and Parthenocarpy in Tomato.Genes (Basel). 2020 Nov 30;11(12):1441. doi: 10.3390/genes11121441. Genes (Basel). 2020. PMID: 33265980 Free PMC article. Review.
Cited by
-
Endogenous Hormone Levels and Transcriptomic Analysis Reveal the Mechanisms of Bulbil Initiation in Pinellia ternata.Int J Mol Sci. 2024 Jun 3;25(11):6149. doi: 10.3390/ijms25116149. Int J Mol Sci. 2024. PMID: 38892337 Free PMC article.
-
Molecular evolution, diversification, and expression assessment of MADS gene family in Setaria italica, Setaria viridis, and Panicum virgatum.Plant Cell Rep. 2023 Jun;42(6):1003-1024. doi: 10.1007/s00299-023-03009-6. Epub 2023 Apr 3. Plant Cell Rep. 2023. PMID: 37012438
-
Genome-wide characterization and expression profiling of MADS-box family genes during organ development and drought stress in Camelina sativa L.Sci Rep. 2025 Mar 18;15(1):9327. doi: 10.1038/s41598-025-93724-9. Sci Rep. 2025. PMID: 40102605 Free PMC article.
-
Genome-wide characterization and expression analysis of MADS-box transcription factor gene family in Perilla frutescens.Front Plant Sci. 2024 Jan 8;14:1299902. doi: 10.3389/fpls.2023.1299902. eCollection 2023. Front Plant Sci. 2024. PMID: 38259943 Free PMC article.
-
Identification of Transcriptional Networks Involved in De Novo Organ Formation in Tomato Hypocotyl Explants.Int J Mol Sci. 2022 Dec 17;23(24):16112. doi: 10.3390/ijms232416112. Int J Mol Sci. 2022. PMID: 36555756 Free PMC article.
References
-
- Alvarezbuylla E.R., Pelaz S., Liljegren S.J., Gold S.E., Burgeff C., Ditta G.S., Ribas d.P.L., Martínezcastilla L., Yanofsky M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA. 2000;97:5328–5333. doi: 10.1073/pnas.97.10.5328. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

