Optimal clustering with missing values
- PMID: 31216989
- PMCID: PMC6584727
- DOI: 10.1186/s12859-019-2832-3
Optimal clustering with missing values
Abstract
Background: Missing values frequently arise in modern biomedical studies due to various reasons, including missing tests or complex profiling technologies for different omics measurements. Missing values can complicate the application of clustering algorithms, whose goals are to group points based on some similarity criterion. A common practice for dealing with missing values in the context of clustering is to first impute the missing values, and then apply the clustering algorithm on the completed data.
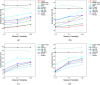
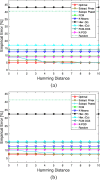
Results: We consider missing values in the context of optimal clustering, which finds an optimal clustering operator with reference to an underlying random labeled point process (RLPP). We show how the missing-value problem fits neatly into the overall framework of optimal clustering by incorporating the missing value mechanism into the random labeled point process and then marginalizing out the missing-value process. In particular, we demonstrate the proposed framework for the Gaussian model with arbitrary covariance structures. Comprehensive experimental studies on both synthetic and real-world RNA-seq data show the superior performance of the proposed optimal clustering with missing values when compared to various clustering approaches.
Conclusion: Optimal clustering with missing values obviates the need for imputation-based pre-processing of the data, while at the same time possessing smaller clustering errors.
Keywords: Clustering; Missing data; Optimal design; Pattern recognition.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Ben-Dor A, Shamir R, Yakhini Z. Clustering gene expression patterns. J Comput Biol. 1999;6:281–97. - PubMed
-
- Bittner M, Meitzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, Sampas N, Dougherty E, Wang E, Marincola F, Gooden C, Lueders J, Glatfelter A, Pollock P, Carpten J, Gillanders E, Leja D, Dietrich K, Beaudry C, Berens M, Alberts D, Sondak V, Hayward N, Trent J. Molecular classification of cutaneous malignant melanoma by gene expression profiling. J Comput Biol. 2000;406(3):536–40. - PubMed
-
- Yeung KY, Fraley C, Murua A, Raftery AE, Ruzzo WL. Model-based clustering and data transformations for gene expression data. Bioinformatics. 2001;17(10):977–87. - PubMed
-
- Fraley C, Raftery AE. Model-based clustering, discriminant analysis, and density estimation. J Am Stat Assoc. 2002;97(458):611–31.
-
- MacEachern SN, Müller P. Estimating mixture of Dirichlet process models. J Comput Graph Stat. 1998;7(2):223–38.
MeSH terms
LinkOut - more resources
Full Text Sources

