Engulfment and cell motility protein 1 potentiates diabetic cardiomyopathy via Rac-dependent and Rac-independent ROS production
- PMID: 31217360
- PMCID: PMC6629098
- DOI: 10.1172/jci.insight.127660
Engulfment and cell motility protein 1 potentiates diabetic cardiomyopathy via Rac-dependent and Rac-independent ROS production
Abstract
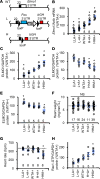
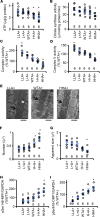
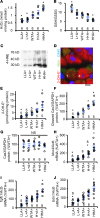
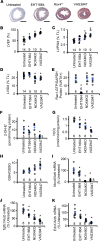
Engulfment and cell motility protein 1 (ELMO1) is part of a guanine nucleotide exchange factor for Ras-related C3 botulinum toxin substrate (Rac), and ELMO1 polymorphisms were identified to be associated with diabetic nephropathy in genome-wide association studies. We generated a set of Akita Ins2C96Y diabetic mice having 5 graded cardiac mRNA levels of ELMO1 from 30% to 200% of normal and found that severe dilated cardiomyopathy develops in ELMO1-hypermorphic mice independent of renal function at age 16 weeks, whereas ELMO1-hypomorphic mice were completely protected. As ELMO1 expression increased, reactive oxygen species indicators, dissociation of the intercalated disc, mitochondrial fragmentation/dysfunction, cleaved caspase-3 levels, and actin polymerization increased in hearts from Akita mice. Cardiomyocyte-specific overexpression in otherwise ELMO1-hypomorphic Akita mice was sufficient to promote cardiomyopathy. Cardiac Rac1 activity was positively correlated with the ELMO1 levels, and oral administration of a pan-Rac inhibitor, EHT1864, partially mitigated cardiomyopathy of the ELMO1 hypermorphs. Disrupting Nox4, a Rac-independent NADPH oxidase, also partially mitigated it. In contrast, a pan-NADPH oxidase inhibitor, VAS3947, markedly prevented cardiomyopathy. Our data demonstrate that in diabetes mellitus ELMO1 is the "rate-limiting" factor of reactive oxygen species production via both Rac-dependent and Rac-independent NADPH oxidases, which in turn trigger cellular signaling cascades toward cardiomyopathy.
Keywords: Cardiology; Cardiovascular disease.
Conflict of interest statement
Figures


Similar articles
-
The bipartite rac1 Guanine nucleotide exchange factor engulfment and cell motility 1/dedicator of cytokinesis 180 (elmo1/dock180) protects endothelial cells from apoptosis in blood vessel development.J Biol Chem. 2015 Mar 6;290(10):6408-18. doi: 10.1074/jbc.M114.633701. Epub 2015 Jan 13. J Biol Chem. 2015. PMID: 25586182 Free PMC article.
-
High Elmo1 expression aggravates and low Elmo1 expression prevents diabetic nephropathy.Proc Natl Acad Sci U S A. 2016 Feb 23;113(8):2218-22. doi: 10.1073/pnas.1600511113. Epub 2016 Feb 8. Proc Natl Acad Sci U S A. 2016. PMID: 26858454 Free PMC article.
-
Elmo1 helps dock180 to regulate Rac1 activity and cell migration of ovarian cancer.Int J Gynecol Cancer. 2014 Jun;24(5):844-50. doi: 10.1097/IGC.0000000000000137. Int J Gynecol Cancer. 2014. PMID: 24819662
-
Production of reactive oxygen species in the diabetic heart. Roles of mitochondria and NADPH oxidase.Circ J. 2014;78(2):300-6. doi: 10.1253/circj.cj-13-1187. Epub 2013 Dec 13. Circ J. 2014. PMID: 24334638 Review.
-
Role of ELMO1 in inflammation and cancer-clinical implications.Cell Oncol (Dordr). 2022 Aug;45(4):505-525. doi: 10.1007/s13402-022-00680-x. Epub 2022 Jun 6. Cell Oncol (Dordr). 2022. PMID: 35668246 Review.
Cited by
-
Cyanocobalamin prevents cardiomyopathy in type 1 diabetes by modulating oxidative stress and DNMT-SOCS1/3-IGF-1 signaling.Commun Biol. 2021 Jun 23;4(1):775. doi: 10.1038/s42003-021-02291-y. Commun Biol. 2021. PMID: 34163008 Free PMC article.
-
Genomic and cellular context-dependent expression of the human ELMO1 gene transcript variants.Gene. 2025 Jun 20;954:149438. doi: 10.1016/j.gene.2025.149438. Epub 2025 Mar 25. Gene. 2025. PMID: 40147730 Free PMC article.
-
Molecular Insights into Oxidative-Stress-Mediated Cardiomyopathy and Potential Therapeutic Strategies.Biomolecules. 2025 May 6;15(5):670. doi: 10.3390/biom15050670. Biomolecules. 2025. PMID: 40427563 Free PMC article. Review.
-
Nox4 as a novel therapeutic target for diabetic vascular complications.Redox Biol. 2023 Aug;64:102781. doi: 10.1016/j.redox.2023.102781. Epub 2023 Jun 9. Redox Biol. 2023. PMID: 37321060 Free PMC article. Review.
-
Islet sympathetic innervation and islet neuropathology in patients with type 1 diabetes.Sci Rep. 2021 Mar 22;11(1):6562. doi: 10.1038/s41598-021-85659-8. Sci Rep. 2021. PMID: 33753784 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

