Comparison of the gut microbiotas of laboratory and wild Asian house shrews (Suncus murinus) based on cloned 16S rRNA sequences
- PMID: 31217361
- PMCID: PMC6842809
- DOI: 10.1538/expanim.19-0021
Comparison of the gut microbiotas of laboratory and wild Asian house shrews (Suncus murinus) based on cloned 16S rRNA sequences
Abstract
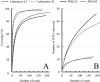
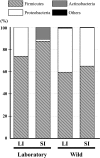
The Asian house shrew, Suncus murinus, is an insectivore (Eulipotyphla, Mammalia) and an important laboratory animal for life-science studies. The gastrointestinal tract of Suncus is simple: the length of the entire intestine is very short relative to body size, the large intestine is quite short, and there are no fermentative chambers such as the forestomach or cecum. These features imply that Suncus has a different nutritional physiology from those of humans and mice, but little is known about whether Suncus utilizes microbial fermentation in the large (LI) or small (SI) intestine. In addition, domestication may affect the gastrointestinal microbial diversity of Suncus. Therefore, we compared the gastrointestinal microbial diversity of Suncus between laboratory and wild Suncus and between the SI and LI (i.e., four groups: Lab-LI, Lab-SI, Wild-LI, and Wild-SI) using bacterial 16S rRNA gene library sequencing analyses with a sub-cloning method. We obtained 759 cloned sequences (176, 174, 195, and 214 from the Lab-LI, Lab-SI, Wild-LI, and Wild-SI samples, respectively), which revealed that the gastrointestinal microbiota of Suncus is rich in Firmicutes (mostly lactic acid bacteria), with few Bacteroidetes. We observed different bacterial communities according to intestinal region in laboratory Suncus, but not in wild Suncus. Furthermore, the gastrointestinal microbial diversity estimates were lower in laboratory Suncus than in wild Suncus. These results imply that Suncus uses lactic acid fermentation in the gut, and that the domestication process altered the gastrointestinal bacterial diversity.
Keywords: Eulipotyphla; domestication; insectivore; microflora; symbiosis.
Figures

References
-
- Chao A., Lee S.M.1992. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87: 210–217. doi: 10.1080/01621459.1992.10475194 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

