The analyses of SRCR genes based on protein-protein interaction network in esophageal squamous cell carcinoma
- PMID: 31217847
- PMCID: PMC6556668
The analyses of SRCR genes based on protein-protein interaction network in esophageal squamous cell carcinoma
Abstract
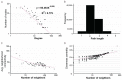
The scavenger receptor cysteine-rich (SRCR) proteins, with one to several SRCR domains, play important roles in human diseases. A full view of their functions in esophageal squamous cell carcinoma (ESCC) remain unclear. Sequence alignment and phylogenetic tree for all human SRCR domains were performed. Differentially-expressed SRCR genes were identified in ESCC, followed by protein-protein interaction (PPI) network construction, topological parameters, subcellular distribution, functional enrichment and survival analyses. The variation of conserved cysteines in each SRCR domain suggested a requirement for new classification of the SRCR family. Six genes (LGALS3BP, MSR1, CD163, LOXL2, LOXL3 and LOXL4) were upregulated, and four genes (DMBT1, PRSS12, TMPRSS2 and SCARA5) were downregulated in ESCC. These 10 SRCR genes form a unique biological network. Functional enrichment analyses provided important clues to investigate the biological functions for SRCR gene network in ESCC, such as extracellular structure organization and the PI3K-Akt signaling pathway. Kaplan-Meier curves confirmed that high expression of SCARA5, LOXL2, LOXL3, LOXL4 were related to poor survival, whereas high expression of DMBTI and PRSS12 showed the opposite result. SRCR genes promote the development of ESCC through its network and could serve as potential prognostic factors and therapy targets of ESCC.
Keywords: SRCR gene; esophageal squamous cell carcinoma; functional enrichment; protein-protein interaction network; sequence analysis.
Conflict of interest statement
None.
Figures






Similar articles
-
Clinical significance of LOXL4 expression and features of LOXL4-associated protein-protein interaction network in esophageal squamous cell carcinoma.Amino Acids. 2019 May;51(5):813-828. doi: 10.1007/s00726-019-02723-4. Epub 2019 Mar 21. Amino Acids. 2019. PMID: 30900087
-
The protein-protein interaction network and clinical significance of heat-shock proteins in esophageal squamous cell carcinoma.Amino Acids. 2018 Jun;50(6):685-697. doi: 10.1007/s00726-018-2569-8. Epub 2018 Apr 27. Amino Acids. 2018. PMID: 29700654
-
Identification and characterization of a cell surface scavenger receptor cysteine-rich protein of Sciaenops ocellatus: bacterial interaction and its dependence on the conserved structural features of the SRCR domain.Fish Shellfish Immunol. 2013 Mar;34(3):810-8. doi: 10.1016/j.fsi.2012.12.016. Epub 2013 Jan 2. Fish Shellfish Immunol. 2013. PMID: 23291106
-
The Scavenger Receptor Cysteine-Rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system.Crit Rev Immunol. 2004;24(1):1-37. doi: 10.1615/critrevimmunol.v24.i1.10. Crit Rev Immunol. 2004. PMID: 14995912 Review.
-
Deleted in malignant brain tumors-1 protein (DMBT1): a pattern recognition receptor with multiple binding sites.Int J Mol Sci. 2010;11(12):5212-33. doi: 10.3390/ijms1112521. Epub 2010 Dec 17. Int J Mol Sci. 2010. PMID: 21614203 Free PMC article. Review.
Cited by
-
The dual role of LOXL4 in the pathogenesis and development of human malignant tumors: a narrative review.Transl Cancer Res. 2024 Apr 30;13(4):2026-2042. doi: 10.21037/tcr-23-2003. Epub 2024 Apr 17. Transl Cancer Res. 2024. PMID: 38737700 Free PMC article. Review.
-
The Expression Pattern and Clinical Significance of Lysyl Oxidase Family in Gliomas.Dokl Biochem Biophys. 2023 Jun;510(1):132-143. doi: 10.1134/S1607672922600269. Epub 2023 Aug 15. Dokl Biochem Biophys. 2023. PMID: 37582875
References
-
- Sarrias MR, Gronlund J, Padilla O, Madsen J, Holmskov U, Lozano F. The Scavenger Receptor Cysteine-Rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit Rev Immunol. 2004;24:1–37. - PubMed
-
- Canton J, Neculai D, Grinstein S. Scavenger receptors in homeostasis and immunity. Nat Rev Immunol. 2013;13:621–634. - PubMed
-
- Martinez VG, Moestrup SK, Holmskov U, Mollenhauer J, Lozano F. The conserved scavenger receptor cysteine-rich superfamily in therapy and diagnosis. Pharmacol Rev. 2011;63:967–1000. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
