miR-23a-5p inhibits cell proliferation and invasion in pancreatic ductal adenocarcinoma by suppressing ECM1 expression
- PMID: 31217868
- PMCID: PMC6556669
miR-23a-5p inhibits cell proliferation and invasion in pancreatic ductal adenocarcinoma by suppressing ECM1 expression
Abstract
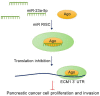
Pancreatic ductal adenocarcinoma (PDAC) is a genetic disease and a leading cause of cancer-related mortality. However, the molecular mechanism underlying PDAC progression remains unclear. In this study, we first confirmed that ECM1 is significantly upregulated in PDAC tissues and that its high levels of expression are closely associated with an advanced histologic grade and a poor prognosis using The Cancer Genome Atlas (TCGA) dataset and the Gene Expression Omnibus (GEO) database. We then found that miR-23a-5p binds directly to the ECM1 3'-untranslated region (3'-UTR), thereby inhibiting ECM1 expression. Functional studies revealed that the induced expression of ECM1 promoted oncogenic abilities and reversed the suppressive effects induced by miR-23a-5p. Collectively, our findings indicate that ECM1 is a proto-oncogene and show that targeting the miR-23a-5p/ECM1 axis may represent a promising therapeutic strategy for PDAC.
Keywords: ECM1; invasion; miR-23a-5p; pancreatic ductal adenocarcinoma (PDAC); proliferation.
Conflict of interest statement
None.
Figures



Similar articles
-
Down-regulation of microRNA-23a promotes pancreatic ductal adenocarcinoma initiation and progression by up-regulation of FOXM1 expression.Genes Dis. 2023 Dec 22;11(5):101203. doi: 10.1016/j.gendis.2023.101203. eCollection 2024 Sep. Genes Dis. 2023. PMID: 39022126 Free PMC article.
-
MicroRNA-769-5p Inhibits Pancreatic Ductal Adenocarcinoma Progression by Directly Targeting and Downregulating ETS Proto-Oncogene 1.Onco Targets Ther. 2019 Dec 31;12:11737-11750. doi: 10.2147/OTT.S218876. eCollection 2019. Onco Targets Ther. 2019. Retraction in: Onco Targets Ther. 2021 Oct 01;14:4965-4966. doi: 10.2147/OTT.S341749. PMID: 32099382 Free PMC article. Retracted.
-
MiRNA-615-5p functions as a tumor suppressor in pancreatic ductal adenocarcinoma by targeting AKT2.PLoS One. 2015 Apr 9;10(4):e0119783. doi: 10.1371/journal.pone.0119783. eCollection 2015. PLoS One. 2015. PMID: 25856297 Free PMC article.
-
miR-23a acts as an oncogene in pancreatic carcinoma by targeting FOXP2.J Investig Med. 2018 Mar;66(3):676-683. doi: 10.1136/jim-2017-000598. Epub 2017 Nov 14. J Investig Med. 2018. PMID: 29141872
-
Circulating microRNAs as Potential Diagnostic, Prognostic and Therapeutic Targets in Pancreatic Cancer.Curr Pharm Des. 2016;22(42):6444-6450. doi: 10.2174/1381612822666160817095047. Curr Pharm Des. 2016. PMID: 27539232 Review.
Cited by
-
Gene Expression Profiles Identified Novel Urine Biomarkers for Diagnosis and Prognosis of High-Grade Bladder Urothelial Carcinoma.Front Oncol. 2020 Mar 27;10:394. doi: 10.3389/fonc.2020.00394. eCollection 2020. Front Oncol. 2020. PMID: 32292720 Free PMC article.
-
MicroRNA-23a-5p regulates cell proliferation, migration and inflammation of TNF-α-stimulated human fibroblast-like MH7A synoviocytes by targeting TLR4 in rheumatoid arthritis.Exp Ther Med. 2021 May;21(5):479. doi: 10.3892/etm.2021.9910. Epub 2021 Mar 12. Exp Ther Med. 2021. PMID: 33767774 Free PMC article.
-
LncRNA FLVCR1-AS1 mediates miR-23a-5p/SLC7A11 axis to promote malignant behavior of cervical cancer cells.Bioengineered. 2022 Apr;13(4):10454-10466. doi: 10.1080/21655979.2022.2059958. Bioengineered. 2022. PMID: 35465835 Free PMC article.
-
Extracellular Matrix Protein 1 Regulates Colorectal Cancer Cell Proliferative, Migratory, Invasive and Epithelial-Mesenchymal Transition Activities Through the PI3K/AKT/GSK3β/Snail Signaling Axis.Front Oncol. 2022 Apr 27;12:889159. doi: 10.3389/fonc.2022.889159. eCollection 2022. Front Oncol. 2022. PMID: 35574325 Free PMC article.
-
Brusatol Inhibits Proliferation and Invasion of Glioblastoma by Down-Regulating the Expression of ECM1.Front Pharmacol. 2021 Dec 14;12:775680. doi: 10.3389/fphar.2021.775680. eCollection 2021. Front Pharmacol. 2021. PMID: 34970146 Free PMC article.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2017;67:7–30. - PubMed
-
- Malvezzi M, Bertuccio P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2014. Ann Oncol. 2014;25:1650–1656. - PubMed
-
- Garrido-Laguna I, Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol. 2015;12:319–334. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
