Functional interplay between YY1 and CARM1 promotes oral carcinogenesis
- PMID: 31217904
- PMCID: PMC6557205
- DOI: 10.18632/oncotarget.26984
Functional interplay between YY1 and CARM1 promotes oral carcinogenesis
Abstract
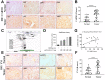
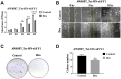
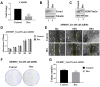
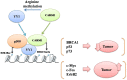
Coactivator associated arginine methyltransferase 1 (CARM1) has been functionally implicated in maintenance of pluripotency, cellular differentiation and tumorigenesis; where it plays regulatory roles by virtue of its ability to coactivate transcription as well as to modulate protein function as an arginine methyltransferase. Previous studies establish an oncogenic function of CARM1 in the context of colorectal and breast cancer, which correlate to its overexpressed condition. However, the mechanism behind its deregulated expression in the context of cancer has not been addressed before. In the present study we uncover an oncogenic function of CARM1 in the context of oral cancer, where it was found to be overexpressed. We also identify YY1 to be a positive regulator of CARM1 gene promoter, where silencing of YY1 in oral cancer cell line could lead to reduction in expression of CARM1. In this context, YY1 showed concomitant overexpression in oral cancer patient samples compared to adjacent normal tissue. Cell line based experiments as well as xenograft study revealed pro-neoplastic functions of YY1 in oral cancer. Transcriptomics analysis as well as qRT-PCR validation clearly indicated pro-proliferative, pro-angiogenic and pro-metastatic role of YY1 in oral cancer. We also show that YY1 is a substrate of CARM1 mediated arginine methylation, where the latter could coactivate YY1 mediated reporter gene activation in vivo. Taken together, CARM1 and YY1 were found to regulate each other in a positive feedback loop to facilitate oral cancer progression.
Keywords: CARM1; YY1; arginine methylation; oncogene; oral cancer.
Conflict of interest statement
CONFLICTS OF INTEREST Authors declare that they have no conflicts of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

