svtools: population-scale analysis of structural variation
- PMID: 31218349
- PMCID: PMC6853660
- DOI: 10.1093/bioinformatics/btz492
svtools: population-scale analysis of structural variation
Abstract
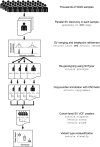
Summary: Large-scale human genetics studies are now employing whole genome sequencing with the goal of conducting comprehensive trait mapping analyses of all forms of genome variation. However, methods for structural variation (SV) analysis have lagged far behind those for smaller scale variants, and there is an urgent need to develop more efficient tools that scale to the size of human populations. Here, we present a fast and highly scalable software toolkit (svtools) and cloud-based pipeline for assembling high quality SV maps-including deletions, duplications, mobile element insertions, inversions and other rearrangements-in many thousands of human genomes. We show that this pipeline achieves similar variant detection performance to established per-sample methods (e.g. LUMPY), while providing fast and affordable joint analysis at the scale of ≥100 000 genomes. These tools will help enable the next generation of human genetics studies.
Availability and implementation: svtools is implemented in Python and freely available (MIT) from https://github.com/hall-lab/svtools.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2019. Published by Oxford University Press.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

