Plasmid profiles among some ESKAPE pathogens in a tertiary care centre in south India
- PMID: 31219087
- PMCID: PMC6563733
- DOI: 10.4103/ijmr.IJMR_2098_17
Plasmid profiles among some ESKAPE pathogens in a tertiary care centre in south India
Abstract
Background & objectives: Plasmid has led to increase in resistant bacterial pathogens through the exchange of antimicrobial resistance (AMR) genetic determinants through horizontal gene transfer. Baseline data on the occurrence of plasmids carrying AMR genes are lacking in India. This study was aimed to identify the plasmids associated with AMR genetic determinants in ESKAPE pathogens.
Methods: A total of 112 ESKAPE isolates including Escherichia coli (n=37), Klebsiella pneumoniae (n=48, including 7 pan-drug susceptible isolates), Acinetobacter baumannii (n=8), Pseudomonas aeruginosa (n=1) and Staphylococcus aureus (n=18) were analyzed in the study. Isolates were screened for antimicrobial susceptibility and whole genome sequencing of isolates was performed using Ion Torrent (PGM) sequencer. Downstream data analysis was done using PATRIC, ResFinder, PlasmidFinder and MLSTFinder databases. All 88 whole genome sequences (WGS) were deposited at GenBank.
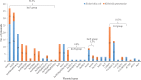
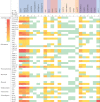
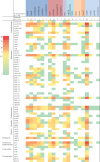
Results: Most of the study isolates showed resistant phenotypes. As analyzed from WGS, the isolates included both known and unknown sequence types. The plasmid analysis revealed the presence of single or multiple plasmids in the isolates. Plasmid types such as IncHI1B(pNDM-MAR), IncFII(pRSB107), IncFIB(Mar), IncFIB(pQil), IncFIA, IncFII(K), IncR, ColKP3 and ColpVC were present in K. pneumoniae. In E. coli, IncFIA, IncFII, IncFIB, Col(BS512), IncL1, IncX3 and IncH were present along with other types. S. aureus harboured seven different plasmid groups pMW2 (rep 5), pSAS1 (rep 7), pDLK1 (rep 10), pUB110 (rep US12), Saa6159 (rep 16), pKH12 (rep 21) and pSA1308 (rep 21). The overall incidence of IncF type plasmids was 56.5 per cent followed by Col type plasmids 18.3 per cent and IncX 5.3 per cent. Other plasmid types identified were <5 per cent.
Interpretation & conclusions: Results from the study may serve as a baseline data for the occurrence of AMR genes and plasmids in India. Information on the association between phenotypic and genotypic expression of AMR was deciphered from the data. Further studies on the mechanism of antibiotic resistance dissemination are essential for enhancing clinical lifetime of antibiotics.
Keywords: Antimicrobial resistance - β; IncF - plasmids; col-horizontal gene transfer; lactamase.
Conflict of interest statement
None
Figures



References
-
- Veeraraghavan B, Jesudason MR, Prakasah JAJ, Anandan S, Sahni RD, Pragasam AK, et al. Antimicrobial susceptibility profiles of Gram-negative bacteria causing infections collected across India during 2014-2016: Study for monitoring antimicrobial resistance trend report. Indian J Med Microbiol. 2018;36:32–6. - PubMed
-
- García-Fernández A, Chiaretto G, Bertini A, Villa L, Fortini D, Ricci A, et al. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum beta-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother. 2008;61:1229–33. - PubMed
-
- Jensen LB, Garcia-Migura L, Valenzuela AJ, Løhr M, Hasman H, Aarestrup FM. A classification system for plasmids from enterococci and other Gram-positive bacteria. J Microbiol Methods. 2010;80:25–43. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
