Association Between Single-Nucleotide Polymorphisms in HLA Alleles and Human Immunodeficiency Virus Type 1 Viral Load in Demographically Diverse, Antiretroviral Therapy-Naive Participants From the Strategic Timing of AntiRetroviral Treatment Trial
- PMID: 31219150
- PMCID: PMC6743845
- DOI: 10.1093/infdis/jiz294
Association Between Single-Nucleotide Polymorphisms in HLA Alleles and Human Immunodeficiency Virus Type 1 Viral Load in Demographically Diverse, Antiretroviral Therapy-Naive Participants From the Strategic Timing of AntiRetroviral Treatment Trial
Abstract
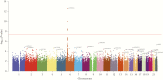
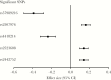
The impact of variation in host genetics on replication of human immunodeficiency virus type 1 (HIV-1) in demographically diverse populations remains uncertain. In the current study, we performed a genome-wide screen for associations of single-nucleotide polymorphisms (SNPs) to viral load (VL) in antiretroviral therapy-naive participants (n = 2440) with varying demographics from the Strategic Timing of AntiRetroviral Treatment (START) trial. Associations were assessed using genotypic data generated by a customized SNP array, imputed HLA alleles, and multiple linear regression. Genome-wide significant associations between SNPs and VL were observed in the major histocompatibility complex class I region (MHC I), with effect sizes ranging between 0.14 and 0.39 log10 VL (copies/mL). Supporting the SNP findings, we identified several HLA alleles significantly associated with VL, extending prior observations that the (MHC I) is a major host determinant of HIV-1 control with shared genetic variants across diverse populations and underscoring the limitations of genome-wide association studies as being merely a screening tool.
Keywords: GWAS; HIV-1; HLA; genome-wide association study; host genetics; viral load.
© The Author(s) 2019. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
Figures

Similar articles
-
Human Immunotypes Impose Selection on Viral Genotypes Through Viral Epitope Specificity.J Infect Dis. 2021 Dec 15;224(12):2053-2063. doi: 10.1093/infdis/jiab253. J Infect Dis. 2021. PMID: 33974707 Free PMC article.
-
Impact of polymorphisms in the HCP5 and HLA-C, and ZNRD1 genes on HIV viral load.Infect Genet Evol. 2016 Jul;41:185-190. doi: 10.1016/j.meegid.2016.03.037. Epub 2016 Apr 12. Infect Genet Evol. 2016. PMID: 27083073
-
A genome-to-genome analysis of associations between human genetic variation, HIV-1 sequence diversity, and viral control.Elife. 2013 Oct 29;2:e01123. doi: 10.7554/eLife.01123. Elife. 2013. PMID: 24171102 Free PMC article.
-
HIV-1 dynamics: a reappraisal of host and viral factors, as well as methodological issues.Viruses. 2012 Oct 10;4(10):2080-96. doi: 10.3390/v4102080. Viruses. 2012. PMID: 23202454 Free PMC article. Review.
-
HIV and HLA class I: an evolving relationship.Immunity. 2012 Sep 21;37(3):426-40. doi: 10.1016/j.immuni.2012.09.005. Immunity. 2012. PMID: 22999948 Free PMC article. Review.
Cited by
-
Human Immunotypes Impose Selection on Viral Genotypes Through Viral Epitope Specificity.J Infect Dis. 2021 Dec 15;224(12):2053-2063. doi: 10.1093/infdis/jiab253. J Infect Dis. 2021. PMID: 33974707 Free PMC article.
-
A year of COVID-19 GWAS results from the GRASP portal reveals potential genetic risk factors.HGG Adv. 2022 Apr 14;3(2):100095. doi: 10.1016/j.xhgg.2022.100095. Epub 2022 Feb 22. HGG Adv. 2022. PMID: 35224516 Free PMC article.
-
Genetic architecture of cardiometabolic risks in people living with HIV.BMC Med. 2020 Oct 28;18(1):288. doi: 10.1186/s12916-020-01762-z. BMC Med. 2020. PMID: 33109212 Free PMC article.
-
Analysis of polygenic selection in purebred and crossbred pig genomes using generation proxy selection mapping.Genet Sel Evol. 2023 Sep 14;55(1):62. doi: 10.1186/s12711-023-00836-9. Genet Sel Evol. 2023. PMID: 37710159 Free PMC article.
-
Current HLA Investigations on SARS-CoV-2 and Perspectives.Front Genet. 2021 Nov 29;12:774922. doi: 10.3389/fgene.2021.774922. eCollection 2021. Front Genet. 2021. PMID: 34912378 Free PMC article. Review.
References
-
- Mellors JW, Rinaldo CR Jr, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 1996; 272:1167–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

