Longitudinal neuroanatomical and cognitive progression of posterior cortical atrophy
- PMID: 31219516
- PMCID: PMC6598737
- DOI: 10.1093/brain/awz136
Longitudinal neuroanatomical and cognitive progression of posterior cortical atrophy
Abstract
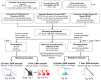
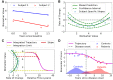
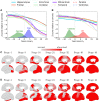
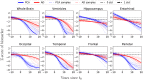
Posterior cortical atrophy is a clinico-radiological syndrome characterized by progressive decline in visual processing and atrophy of posterior brain regions. With the majority of cases attributable to Alzheimer's disease and recent evidence for genetic risk factors specifically related to posterior cortical atrophy, the syndrome can provide important insights into selective vulnerability and phenotypic diversity. The present study describes the first major longitudinal investigation of posterior cortical atrophy disease progression. Three hundred and sixty-one individuals (117 posterior cortical atrophy, 106 typical Alzheimer's disease, 138 controls) fulfilling consensus criteria for posterior cortical atrophy-pure and typical Alzheimer's disease were recruited from three centres in the UK, Spain and USA. Participants underwent up to six annual assessments involving MRI scans and neuropsychological testing. We constructed longitudinal trajectories of regional brain volumes within posterior cortical atrophy and typical Alzheimer's disease using differential equation models. We compared and contrasted the order in which regional brain volumes become abnormal within posterior cortical atrophy and typical Alzheimer's disease using event-based models. We also examined trajectories of cognitive decline and the order in which different cognitive tests show abnormality using the same models. Temporally aligned trajectories for eight regions of interest revealed distinct (P < 0.002) patterns of progression in posterior cortical atrophy and typical Alzheimer's disease. Patients with posterior cortical atrophy showed early occipital and parietal atrophy, with subsequent higher rates of temporal atrophy and ventricular expansion leading to tissue loss of comparable extent later. Hippocampal, entorhinal and frontal regions underwent a lower rate of change and never approached the extent of posterior cortical involvement. Patients with typical Alzheimer's disease showed early hippocampal atrophy, with subsequent higher rates of temporal atrophy and ventricular expansion. Cognitive models showed tests sensitive to visuospatial dysfunction declined earlier in posterior cortical atrophy than typical Alzheimer's disease whilst tests sensitive to working memory impairment declined earlier in typical Alzheimer's disease than posterior cortical atrophy. These findings indicate that posterior cortical atrophy and typical Alzheimer's disease have distinct sites of onset and different profiles of spatial and temporal progression. The ordering of disease events both motivates investigation of biological factors underpinning phenotypic heterogeneity, and informs the selection of measures for clinical trials in posterior cortical atrophy.
Keywords: Alzheimer’s disease; brain atrophy; dementia; memory; structural MRI.
© The Author(s) (2019). Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures



References
-
- Benson DF, Davis RJ, Snyder BD. Posterior cortical atrophy. Arch Neurol 1988; 45: 789–93. - PubMed
-
- Cardoso MJ, Wolz R, Modat M, Fox NC, Rueckert D, Ourselin S. Geodesic information flows. Med Image Comput Comput-Assist Interv MICCAI Int Conf Med Image Comput Comput-Assist Interv 2012; 15: 262–70. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

