Activation of Wnt signaling promotes olaparib resistant ovarian cancer
- PMID: 31219654
- PMCID: PMC7537108
- DOI: 10.1002/mc.23064
Activation of Wnt signaling promotes olaparib resistant ovarian cancer
Abstract
Epithelial ovarian cancer (EOC) has one of the highest death to incidence ratios among all cancers. High grade serous ovarian carcinoma (HGSOC) is the most common and deadliest EOC histotype due to the lack of therapeutic options following debulking surgery and platinum/taxane-based chemotherapies. For recurrent chemosensitive HGSOC, poly(ADP)-ribose polymerase inhibitors (PARPi; olaparib, rucaparib, or niraparib) represent an emerging treatment strategy. While PARPi are most effective in homologous recombination DNA repair-deficient (HRD) HGSOCs, recent studies have observed a significant benefit in non-HRD HGSOCs. However, all HGSOC patients are likely to acquire resistance. Therefore, there is an urgent clinical need to understand PARPi resistance and to introduce novel combinatorial therapies to manage PARPi resistance and extend HGSOC disease-free intervals. In a panel of HGSOC cell lines, we established matched olaparib sensitive and resistant cells. Transcriptome analysis of the matched olaparib-sensitive vs -resistant cells revealed activation of the Wnt signaling pathway and consequently increased TCF transcriptional activity in PARPi-resistant cells. Forced activation of canonical Wnt signaling in several PARPi-sensitive cells via WNT3A reduced olaparib and rucaparib sensitivity. PARPi resistant cells were sensitive to inhibition of Wnt signaling using the FDA-approved compound, pyrvinium pamoate, which has been shown to promote downregulation of β-catenin. In both an HGSOC cell line and a patient-derived xenograft model, we observed that combining pyrvinium pamoate with olaparib resulted in a significant decrease in tumor burden. This study demonstrates that Wnt signaling can mediate PARPi resistance in HGSOC and provides a clinical rationale for combining PARP and Wnt inhibitors.
Keywords: DNA repair; PARP inhibitor; ovarian cancer; therapy resistance; wnt signaling.
© 2019 Wiley Periodicals, Inc.
Conflict of interest statement
Figures
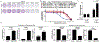

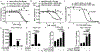

Similar articles
-
N6-Methylation of Adenosine of FZD10 mRNA Contributes to PARP Inhibitor Resistance.Cancer Res. 2019 Jun 1;79(11):2812-2820. doi: 10.1158/0008-5472.CAN-18-3592. Epub 2019 Apr 9. Cancer Res. 2019. PMID: 30967398 Free PMC article.
-
Histone methyltransferases EHMT1 and EHMT2 (GLP/G9A) maintain PARP inhibitor resistance in high-grade serous ovarian carcinoma.Clin Epigenetics. 2019 Nov 27;11(1):165. doi: 10.1186/s13148-019-0758-2. Clin Epigenetics. 2019. PMID: 31775874 Free PMC article.
-
ALDH1A1 Contributes to PARP Inhibitor Resistance via Enhancing DNA Repair in BRCA2-/- Ovarian Cancer Cells.Mol Cancer Ther. 2020 Jan;19(1):199-210. doi: 10.1158/1535-7163.MCT-19-0242. Epub 2019 Sep 18. Mol Cancer Ther. 2020. PMID: 31534014 Free PMC article.
-
The promising PARP inhibitors in ovarian cancer therapy: From Olaparib to others.Biomed Pharmacother. 2018 Mar;99:552-560. doi: 10.1016/j.biopha.2018.01.094. Epub 2018 Feb 20. Biomed Pharmacother. 2018. PMID: 29895102 Review.
-
Cediranib, a pan-VEGFR inhibitor, and olaparib, a PARP inhibitor, in combination therapy for high grade serous ovarian cancer.Expert Opin Investig Drugs. 2016;25(5):597-611. doi: 10.1517/13543784.2016.1156857. Epub 2016 Mar 16. Expert Opin Investig Drugs. 2016. PMID: 26899229 Review.
Cited by
-
WNT4 Regulates Cellular Metabolism via Intracellular Activity at the Mitochondria in Breast and Gynecologic Cancers.Cancer Res Commun. 2024 Jan 17;4(1):134-151. doi: 10.1158/2767-9764.CRC-23-0275. Cancer Res Commun. 2024. PMID: 38112643 Free PMC article.
-
Loss of Claudin-4 Reduces DNA Damage Repair and Increases Sensitivity to PARP Inhibitors.Mol Cancer Ther. 2022 Apr 1;21(4):647-657. doi: 10.1158/1535-7163.MCT-21-0827. Mol Cancer Ther. 2022. PMID: 35373300 Free PMC article.
-
Targeting Fatty Acid Oxidation to Promote Anoikis and Inhibit Ovarian Cancer Progression.Mol Cancer Res. 2020 Jul;18(7):1088-1098. doi: 10.1158/1541-7786.MCR-19-1057. Epub 2020 Mar 20. Mol Cancer Res. 2020. PMID: 32198139 Free PMC article.
-
Cancer stem cells, epithelial-mesenchymal transition, ATP and their roles in drug resistance in cancer.Cancer Drug Resist. 2021;4(3):684-709. doi: 10.20517/cdr.2021.32. Epub 2021 Jun 17. Cancer Drug Resist. 2021. PMID: 34322664 Free PMC article.
-
Downregulation of TAB182 promotes cancer stem-like cell properties and therapeutic resistance in triple-negative breast cancer cells.BMC Cancer. 2023 Nov 13;23(1):1101. doi: 10.1186/s12885-023-11552-4. BMC Cancer. 2023. PMID: 37953246 Free PMC article.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30 - PubMed
-
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917–21 - PubMed
-
- McCabe N, Lord CJ, Tutt AN, Martin NM, Smith GC, Ashworth A. BRCA2-deficient CAPAN-1 cells are extremely sensitive to the inhibition of Poly (ADP-Ribose) polymerase: an issue of potency. Cancer Biol Ther 2005;4:934–6 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

