Response to comment on 'Valid molecular dynamics simulations of human hemoglobin require a surprisingly large box size'
- PMID: 31219783
- PMCID: PMC6586459
- DOI: 10.7554/eLife.45318
Response to comment on 'Valid molecular dynamics simulations of human hemoglobin require a surprisingly large box size'
Abstract
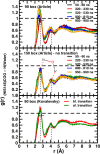
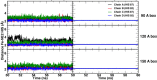
We recently reported that molecular dynamics simulations for hemoglobin require a surprisingly large box size to stabilize the T(0) state relative to R(0), as observed in experiments (El Hage et al., 2018). Gapsys and de Groot have commented on this work but do not provide convincing evidence that the conclusions of El Hage et al., 2018 are incorrect. Here we respond to these concerns, argue that our original conclusions remain valid, and raise our own concerns about some of the results reported in the comment by Gapsys and de Groot that require clarification.
Keywords: box size; computational biology; hemoglobin; hydrophobic effect; molecular biophysics; molecular dynamics; none; structural biology; systems biology.
© 2019, El Hage et al.
Conflict of interest statement
KE, FH, PG, MM, MK No competing interests declared
Figures
Comment on
-
Valid molecular dynamics simulations of human hemoglobin require a surprisingly large box size.Elife. 2018 Jul 12;7:e35560. doi: 10.7554/eLife.35560. Elife. 2018. PMID: 29998846 Free PMC article.
-
Comment on 'Valid molecular dynamics simulations of human hemoglobin require a surprisingly large box size'.Elife. 2019 Jun 20;8:e44718. doi: 10.7554/eLife.44718. Elife. 2019. PMID: 31219782 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

