Protein ensembles link genotype to phenotype
- PMID: 31220071
- PMCID: PMC6586255
- DOI: 10.1371/journal.pcbi.1006648
Protein ensembles link genotype to phenotype
Abstract
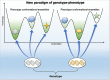
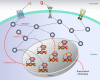
Classically, phenotype is what is observed, and genotype is the genetic makeup. Statistical studies aim to project phenotypic likelihoods of genotypic patterns. The traditional genotype-to-phenotype theory embraces the view that the encoded protein shape together with gene expression level largely determines the resulting phenotypic trait. Here, we point out that the molecular biology revolution at the turn of the century explained that the gene encodes not one but ensembles of conformations, which in turn spell all possible gene-associated phenotypes. The significance of a dynamic ensemble view is in understanding the linkage between genetic change and the gained observable physical or biochemical characteristics. Thus, despite the transformative shift in our understanding of the basis of protein structure and function, the literature still commonly relates to the classical genotype-phenotype paradigm. This is important because an ensemble view clarifies how even seemingly small genetic alterations can lead to pleiotropic traits in adaptive evolution and in disease, why cellular pathways can be modified in monogenic and polygenic traits, and how the environment may tweak protein function.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Frauenfelder H, Sligar SG, Wolynes PG (1991) The energy landscapes and motions of proteins. Science 254(5038):1598–1603. https://10.1126/science.1749933 - DOI - PubMed
-
- Ma B, Kumar S, Tsai CJ, Nussinov R (1999) Folding funnels and binding mechanisms. Protein Eng 12(9):713–720. https://10.1093/protein/12.9.713 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

