Emergence of phylogenetically diverse and fluoroquinolone resistant Salmonella Enteritidis as a cause of invasive nontyphoidal Salmonella disease in Ghana
- PMID: 31220112
- PMCID: PMC6605661
- DOI: 10.1371/journal.pntd.0007485
Emergence of phylogenetically diverse and fluoroquinolone resistant Salmonella Enteritidis as a cause of invasive nontyphoidal Salmonella disease in Ghana
Abstract
Background: Salmonella enterica serovar Enteritidis is a cause of both poultry- and egg-associated enterocolitis globally and bloodstream-invasive nontyphoidal Salmonella (iNTS) disease in sub-Saharan Africa (sSA). Distinct, multi-drug resistant genotypes associated with iNTS disease in sSA have recently been described, often requiring treatment with fluoroquinolone antibiotics. In industrialised countries, antimicrobial use in poultry production has led to frequent fluoroquinolone resistance amongst globally prevalent enterocolitis-associated lineages.
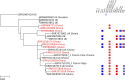
Methodology/principal findings: Twenty seven S. Enteritidis isolates from patients with iNTS disease and two poultry isolates, collected between 2007 and 2015 in the Ashanti region of Ghana, were whole-genome sequenced. These isolates, notable for a high rate of diminished ciprofloxacin susceptibility (DCS), were placed in the phyletic context of 1,067 sequences from the Public Health England (PHE) S. Enteritidis genome database to understand whether DCS was associated with African or globally-circulating clades of S. Enteritidis. Analysis showed four of the major S. Enteritidis clades were represented, two global and two African. All thirteen DCS isolates, containing a single gyrA mutation at codon 87, belonged to a global PT4-like clade responsible for epidemics of poultry-associated enterocolitis. Apart from two DCS isolates, which clustered with PHE isolates associated with travel to Spain and Brazil, the remaining DCS isolates, including one poultry isolate, belonged to two monophyletic clusters in which gyrA 87 mutations appear to have developed within the region.
Conclusions/significance: Extensive phylogenetic diversity is evident amongst iNTS disease-associated S. Enteritidis in Ghana. Antimicrobial resistance profiles differed by clade, highlighting the challenges of devising empirical sepsis guidelines. The detection of fluoroquinolone resistance in phyletically-related poultry and human isolates is of major concern and surveillance and control measures within the region's burgeoning poultry industry are required to protect a human population at high risk of iNTS disease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

