The large genome size variation in the Hesperis clade was shaped by the prevalent proliferation of DNA repeats and rarer genome downsizing
- PMID: 31220201
- PMCID: PMC6676390
- DOI: 10.1093/aob/mcz036
The large genome size variation in the Hesperis clade was shaped by the prevalent proliferation of DNA repeats and rarer genome downsizing
Abstract
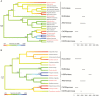
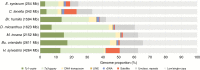
Background and aims: Most crucifer species (Brassicaceae) have small nuclear genomes (mean 1C-value 617 Mb). The species with the largest genomes occur within the monophyletic Hesperis clade (Mandáková et al., Plant Physiology174: 2062-2071; also known as Clade E or Lineage III). Whereas most chromosome numbers in the clade are 6 or 7, monoploid genome sizes vary 16-fold (256-4264 Mb). To get an insight into genome size evolution in the Hesperis clade (~350 species in ~48 genera), we aimed to identify, quantify and localize in situ the repeats from which these genomes are built. We analysed nuclear repeatomes in seven species, covering the phylogenetic and genome size breadth of the clade, by low-pass whole-genome sequencing.
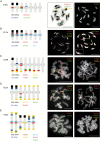
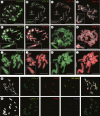
Methods: Genome size was estimated by flow cytometry. Genomic DNA was sequenced on an Illumina sequencer and DNA repeats were identified and quantified using RepeatExplorer; the most abundant repeats were localized on chromosomes by fluorescence in situ hybridization. To evaluate the feasibility of bacterial artificial chromosome (BAC)-based comparative chromosome painting in Hesperis-clade species, BACs of arabidopsis were used as painting probes.
Key results: Most biennial and perennial species of the Hesperis clade possess unusually large nuclear genomes due to the proliferation of long terminal repeat retrotransposons. The prevalent genome expansion was rarely, but repeatedly, counteracted by purging of transposable elements in ephemeral and annual species.
Conclusions: The most common ancestor of the Hesperis clade has experienced genome upsizing due to transposable element amplification. Further genome size increases, dominating diversification of all Hesperis-clade tribes, contrast with the overall stability of chromosome numbers. In some subclades and species genome downsizing occurred, presumably as an adaptive transition to an annual life cycle. The amplification versus purging of transposable elements and tandem repeats impacted the chromosomal architecture of the Hesperis-clade species.
Keywords: Bunias; Hesperis; Matthiola; Brassicaceae; Genome size evolution; Lineage III; chromosome organization; interstitial telomeric repeats (ITRs); repetitive DNA; retrotransposons; tandem repeats.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Annals of Botany Company. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. - PubMed
-
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
-
- Beilstein MA, Al-Shehbaz IA, Kellogg EA. 2006. Brassicaceae phylogeny and trichome evolution. American Journal of Botany 93: 607–619. - PubMed
-
- Bennett MD. 1987. Variation in genomic form in plants and its ecological implications. New Phytologist 106: 177–200.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

