The FUS-DDIT3 Interactome in Myxoid Liposarcoma
- PMID: 31220736
- PMCID: PMC6584455
- DOI: 10.1016/j.neo.2019.05.004
The FUS-DDIT3 Interactome in Myxoid Liposarcoma
Abstract
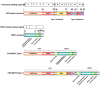
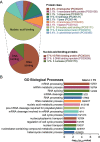
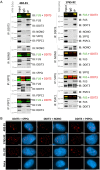
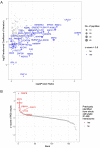
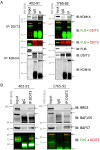
Myxoid liposarcoma is a malignant lipogenic tumor that develops in deep soft tissues. While local control rates are good, current chemotherapy options remain ineffective against metastatic disease. Myxoid liposarcoma is characterized by the FUS-DDIT3 fusion oncoprotein that is proposed to function as an aberrant transcription factor, but its exact mechanism of action has remained unclear. To identify the key functional interacting partners of FUS-DDIT3, this study utilized immunoprecipitation-mass spectrometry (IP-MS) to identify the FUS-DDIT3 interactome in whole cell lysates of myxoid liposarcoma cells, and results showed an enrichment of RNA processing proteins. Further quantitative MS analyses of FUS-DDIT3 complexes isolated from nuclear lysates showed that members of several chromatin regulatory complexes were present in the FUS-DDIT3 interactome, including NuRD, SWI/SNF, PRC1, PRC2, and MLL1 COMPASS-like complexes. Co-immunoprecipitation validated the associations of FUS-DDIT3 with BRG1/SMARCA4, BAF155/SMARCC1, BAF57/SMARCE1, and KDM1A. Data from this study provides candidates for functional validation as potential therapeutic targets, particularly for emerging epigenetic drugs.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Regulatory mechanisms, expression levels and proliferation effects of the FUS-DDIT3 fusion oncogene in liposarcoma.J Pathol. 2016 Apr;238(5):689-99. doi: 10.1002/path.4700. J Pathol. 2016. PMID: 26865464
-
FUS-DDIT3 Fusion Protein-Driven IGF-IR Signaling is a Therapeutic Target in Myxoid Liposarcoma.Clin Cancer Res. 2017 Oct 15;23(20):6227-6238. doi: 10.1158/1078-0432.CCR-17-0130. Epub 2017 Jun 21. Clin Cancer Res. 2017. PMID: 28637688
-
The myxoid liposarcoma FUS-DDIT3 fusion oncoprotein deregulates NF-kappaB target genes by interaction with NFKBIZ.Oncogene. 2009 Jan 15;28(2):270-8. doi: 10.1038/onc.2008.378. Epub 2008 Oct 13. Oncogene. 2009. PMID: 18850010
-
FUS::DDIT3 Fusion Protein in the Development of Myxoid Liposarcoma and Possible Implications for Therapy.Biomolecules. 2024 Oct 14;14(10):1297. doi: 10.3390/biom14101297. Biomolecules. 2024. PMID: 39456230 Free PMC article. Review.
-
Gene of the month: DDIT3.J Clin Pathol. 2024 Mar 20;77(4):211-216. doi: 10.1136/jcp-2023-208963. J Clin Pathol. 2024. PMID: 38053287 Review.
Cited by
-
FUS oncofusion protein condensates recruit mSWI/SNF chromatin remodeler via heterotypic interactions between prion-like domains.Protein Sci. 2021 Jul;30(7):1454-1466. doi: 10.1002/pro.4127. Epub 2021 Jun 4. Protein Sci. 2021. PMID: 34018649 Free PMC article.
-
[Clinical value of fluorescence in situ hybridization with MDM2 and DDIT3 probe in diagnosis of liposarcoma].Beijing Da Xue Xue Bao Yi Xue Ban. 2023 Apr 18;55(2):228-233. doi: 10.19723/j.issn.1671-167X.2023.02.005. Beijing Da Xue Xue Bao Yi Xue Ban. 2023. PMID: 37042132 Free PMC article. Chinese.
-
Survival of a patient with multiple-recurrent giant retroperitoneal dedifferentiated liposarcoma for 15 years: A case report.Front Surg. 2022 Nov 7;9:916802. doi: 10.3389/fsurg.2022.916802. eCollection 2022. Front Surg. 2022. PMID: 36420404 Free PMC article.
-
The Immune Contexture of Liposarcoma and Its Clinical Implications.Cancers (Basel). 2022 Sep 21;14(19):4578. doi: 10.3390/cancers14194578. Cancers (Basel). 2022. PMID: 36230502 Free PMC article. Review.
-
The oncogenic transcription factor FUS-CHOP can undergo nuclear liquid-liquid phase separation.J Cell Sci. 2021 Sep 1;134(17):jcs258578. doi: 10.1242/jcs.258578. Epub 2021 Sep 3. J Cell Sci. 2021. PMID: 34357401 Free PMC article.
References
-
- Aman P, Ron D, Mandahl N, Fioretos T, Heim S, Arheden K, Willén H, Rydholm A, Mitelman F. Rearrangement of the transcription factor gene CHOP in myxoid liposarcomas with t(12;16)(q13;p11) Genes Chromosom Cancer. 1992;5(4):278–285. - PubMed
-
- Panagopoulos I, Höglund M, Mertens F, Mandahl N, Mitelman F, Aman P. Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene. 1996;12(3):489–494. - PubMed
-
- Powers MP, Wang W-L, Hernandez VS, Patel KS, Lev DC, Lazar AJ, López-Terrada DH. Detection of myxoid liposarcoma-associated FUS-DDIT3 rearrangement variants including a newly identified breakpoint using an optimized RT-PCR assay. Mod Pathol. 2010;23(10):1307–1315. - PubMed
-
- Dormann D, Haass C. Fused in sarcoma (FUS): an oncogene goes awry in neurodegeneration. Mol Cell Neurosci. 2013;56:475–486. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

