MLKS2 is an ARM domain and F-actin-associated KASH protein that functions in stomatal complex development and meiotic chromosome segregation
- PMID: 31221013
- PMCID: PMC6649574
- DOI: 10.1080/19491034.2019.1629795
MLKS2 is an ARM domain and F-actin-associated KASH protein that functions in stomatal complex development and meiotic chromosome segregation
Abstract
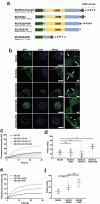
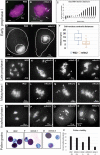
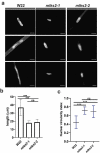
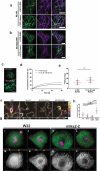
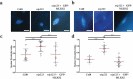
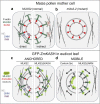
The linker of nucleoskeleton and cytoskeleton (LINC) complex is an essential multi-protein structure spanning the eukaryotic nuclear envelope. The LINC complex functions to maintain nuclear architecture, positioning, and mobility, along with specialized functions in meiotic prophase and chromosome segregation. Members of the LINC complex were recently identified in maize, an important scientific and agricultural grass species. Here we characterized Maize LINC KASH AtSINE-like2, MLKS2, which encodes a highly conserved SINE-group plant KASH protein with characteristic N-terminal armadillo repeats (ARM). Using a heterologous expression system, we showed that actively expressed GFP-MLKS2 is targeted to the nuclear periphery and colocalizes with F-actin and the endoplasmic reticulum, but not microtubules in the cell cortex. Expression of GFP-MLKS2, but not GFP-MLKS2ΔARM, resulted in nuclear anchoring. Genetic analysis of transposon-insertion mutations, mlks2-1 and mlks2-2, showed that the mutant phenotypes were pleiotropic, affecting root hair nuclear morphology, stomatal complex development, multiple aspects of meiosis, and pollen viability. In male meiosis, the mutants showed defects for bouquet-stage telomere clustering, nuclear repositioning, perinuclear actin accumulation, dispersal of late prophase bivalents, and meiotic chromosome segregation. These findings support a model in which the nucleus is connected to cytoskeletal F-actin through the ARM-domain, predicted alpha solenoid structure of MLKS2. Functional conservation of MLKS2 was demonstrated through genetic rescue of the misshapen nuclear phenotype of an Arabidopsis (triple-WIP) KASH mutant. This study establishes a role for the SINE-type KASH proteins in affecting the dynamic nuclear phenomena required for normal plant growth and fertility. Abbreviations: FRAP: Fluorescence recovery after photobleaching; DPI: Days post infiltration; OD: Optical density; MLKS2: Maize LINC KASH AtSINE-like2; LINC: Linker of nucleoskeleton and cytoskeleton; NE: Nuclear envelope; INM: Inner nuclear membrane; ONM: Outer nuclear membrane.
Keywords: FRAP; KASH; LINC; MLKS2; actin; bouquet; maize; meiosis; nuclear envelope; telomere.
Figures



Similar articles
-
The missing LINC: a mammalian KASH-domain protein coupling meiotic chromosomes to the cytoskeleton.Nucleus. 2014 Jan-Feb;5(1):3-10. doi: 10.4161/nucl.27819. Epub 2014 Jan 23. Nucleus. 2014. PMID: 24637401 Free PMC article.
-
Identification and characterization of genes encoding the nuclear envelope LINC complex in the monocot species Zea mays.J Cell Sci. 2019 Feb 11;132(3):jcs221390. doi: 10.1242/jcs.221390. J Cell Sci. 2019. PMID: 30659121
-
A dynamic meiotic SUN belt includes the zygotene-stage telomere bouquet and is disrupted in chromosome segregation mutants of maize (Zea mays L.).Front Plant Sci. 2014 Jul 11;5:314. doi: 10.3389/fpls.2014.00314. eCollection 2014. Front Plant Sci. 2014. PMID: 25071797 Free PMC article.
-
How plants LINC the SUN to KASH.Nucleus. 2013 May-Jun;4(3):206-15. doi: 10.4161/nucl.24088. Epub 2013 May 13. Nucleus. 2013. PMID: 23680964 Free PMC article. Review.
-
The plant LINC complex at the nuclear envelope.Chromosome Res. 2014 Jun;22(2):241-52. doi: 10.1007/s10577-014-9419-7. Chromosome Res. 2014. PMID: 24801343 Review.
Cited by
-
Is the nucleus the unwitting architect of asymmetric cell division in plants?Plant Physiol. 2023 Aug 31;193(1):6-8. doi: 10.1093/plphys/kiad379. Plant Physiol. 2023. PMID: 37399225 Free PMC article. No abstract available.
-
A glossary of plant cell structures: Current insights and future questions.Plant Cell. 2022 Jan 20;34(1):10-52. doi: 10.1093/plcell/koab247. Plant Cell. 2022. PMID: 34633455 Free PMC article. Review.
-
Regulation and Physiological Significance of the Nuclear Shape in Plants.Front Plant Sci. 2021 Jun 10;12:673905. doi: 10.3389/fpls.2021.673905. eCollection 2021. Front Plant Sci. 2021. PMID: 34177991 Free PMC article. Review.
-
Diversity and structural-functional insights of alpha-solenoid proteins.Protein Sci. 2024 Nov;33(11):e5189. doi: 10.1002/pro.5189. Protein Sci. 2024. PMID: 39465903 Free PMC article. Review.
-
Recent advances in understanding the biological roles of the plant nuclear envelope.Nucleus. 2020 Dec;11(1):330-346. doi: 10.1080/19491034.2020.1846836. Nucleus. 2020. PMID: 33161800 Free PMC article.
References
-
- Burke B, Roux KJ. Nuclei take a position: managing nuclear location. Dev Cell. 2009;17:587–597. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
