The hydroxymethylome of multiple myeloma identifies FAM72D as a 1q21 marker linked to proliferation
- PMID: 31221779
- PMCID: PMC7049362
- DOI: 10.3324/haematol.2019.222133
The hydroxymethylome of multiple myeloma identifies FAM72D as a 1q21 marker linked to proliferation
Abstract
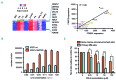
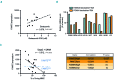
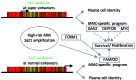
Cell identity relies on the cross-talk between genetics and epigenetics and their impact on gene expression. Oxidation of 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC) is the first step of an active DNA demethylation process occurring mainly at enhancers and gene bodies and, as such, participates in processes governing cell identity in normal and pathological conditions. Although genetic alterations are well documented in multiple myeloma (MM), epigenetic alterations associated with this disease have not yet been thoroughly analyzed. To gain insight into the biology of MM, genome-wide 5hmC profiles were obtained and showed that regions enriched in this modified base overlap with MM enhancers and super enhancers and are close to highly expressed genes. Through the definition of a MM-specific 5hmC signature, we identified FAM72D as a poor prognostic gene located on 1q21, a region amplified in high risk myeloma. We further uncovered that FAM72D functions as part of the FOXM1 transcription factor network controlling cell proliferation and survival and we evidenced an increased sensitivity of cells expressing high levels of FOXM1 and FAM72 to epigenetic drugs targeting histone deacetylases and DNA methyltransferases.
Copyright© 2020 Ferrata Storti Foundation.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

