Microhomology-mediated end joining drives complex rearrangements and overexpression of MYC and PVT1 in multiple myeloma
- PMID: 31221783
- PMCID: PMC7109748
- DOI: 10.3324/haematol.2019.217927
Microhomology-mediated end joining drives complex rearrangements and overexpression of MYC and PVT1 in multiple myeloma
Abstract
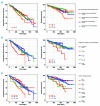
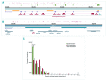
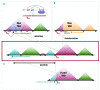
MYC is a widely acting transcription factor and its deregulation is a crucial event in many human cancers. MYC is important biologically and clinically in multiple myeloma, but the mechanisms underlying its dysregulation are poorly understood. We show that MYC rearrangements are present in 36.0% of newly diagnosed myeloma patients, as detected in the largest set of next generation sequencing data to date (n=1,267). Rearrangements were complex and associated with increased expression of MYC and PVT1, but not other genes at 8q24. The highest effect on gene expression was detected in cases where the MYC locus is juxtaposed next to super-enhancers associated with genes such as IGH, IGK, IGL, TXNDC5/BMP6, FAM46C and FOXO3 We identified three hotspots of recombination at 8q24, one of which is enriched for IGH-MYC translocations. Breakpoint analysis indicates primary myeloma rearrangements involving the IGH locus occur through non-homologous end joining, whereas secondary MYC rearrangements occur through microhomology-mediated end joining. This mechanism is different to lymphomas, where non-homologous end joining generates MYC rearrangements. Rearrangements resulted in overexpression of key genes and chromatin immunoprecipitation-sequencing identified that HK2, a member of the glucose metabolism pathway, is directly over-expressed through binding of MYC at its promoter.
Copyright© 2020 Ferrata Storti Foundation.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

