Non-equilibrium dynamics of a nascent polypeptide during translation suppress its misfolding
- PMID: 31221966
- PMCID: PMC6586675
- DOI: 10.1038/s41467-019-10647-6
Non-equilibrium dynamics of a nascent polypeptide during translation suppress its misfolding
Abstract
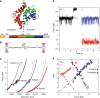
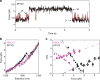
Protein folding can begin co-translationally. Due to the difference in timescale between folding and synthesis, co-translational folding is thought to occur at equilibrium for fast-folding domains. In this scenario, the folding kinetics of stalled ribosome-bound nascent chains should match the folding of nascent chains in real time. To test if this assumption is true, we compare the folding of a ribosome-bound, multi-domain calcium-binding protein stalled at different points in translation with the nascent chain as is it being synthesized in real-time, via optical tweezers. On stalled ribosomes, a misfolded state forms rapidly (1.5 s). However, during translation, this state is only attained after a long delay (63 s), indicating that, unexpectedly, the growing polypeptide is not equilibrated with its ensemble of accessible conformations. Slow equilibration on the ribosome can delay premature folding until adequate sequence is available and/or allow time for chaperone binding, thus promoting productive folding.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM032543/GM/NIGMS NIH HHS/United States
- R01 GM071552/GM/NIGMS NIH HHS/United States
- R01GM071552/U.S. Department of Health & Human Services | National Institutes of Health (NIH)/International
- R01GM032543/U.S. Department of Health & Human Services | National Institutes of Health (NIH)/International
LinkOut - more resources
Full Text Sources
Research Materials

