Genome-wide alternative splicing landscapes modulated by biotrophic sugarcane smut pathogen
- PMID: 31222001
- PMCID: PMC6586842
- DOI: 10.1038/s41598-019-45184-1
Genome-wide alternative splicing landscapes modulated by biotrophic sugarcane smut pathogen
Abstract
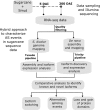
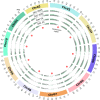
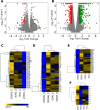
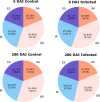
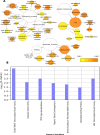
Alternative splicing (AS) promotes transcriptome and proteome diversity during growth, development, and stress responses in eukaryotes. Genome-wide studies of AS in sugarcane (Saccharum spp.) are lacking, mainly due to the absence of a high-quality sequenced reference genome, sugarcane's large, complex genome, and the variable chromosome numbers and polyploidy of sugarcane cultivars. Here, we analyzed changes in the sugarcane isoform-level transcriptome and AS landscape during infection with the smut fungus (Sporisorium scitamineum) using a hybrid approach involving Sorghum bicolor reference-based and Trinity de novo mapping tools. In total, this analysis detected 16,039 and 15,379 transcripts (≥2 FPKM) at 5 and 200 days after infection, respectively. A conservative estimate of isoform-level expression suggested that approximately 5,000 (14%) sugarcane genes undergo AS. Differential expression analysis of the alternatively spliced genes in healthy and smut-infected sugarcane revealed 896 AS events modulated at different stages of infection. Gene family and gene ontology functional enrichment analysis of the differentially spliced genes revealed overrepresentation of functional categories related to the cell wall, defense, and redox homeostasis pathways. Our study provides novel insight into the AS landscape of sugarcane during smut disease interactions.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- FAO. (Latest update: 28 May, 2018. Accessed: 30 Nov, 2018. http://www.fao.org/faostat/en/#data/QC/visualize, 2018).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

