A Hybrid Gene Selection Method Based on ReliefF and Ant Colony Optimization Algorithm for Tumor Classification
- PMID: 31222027
- PMCID: PMC6586811
- DOI: 10.1038/s41598-019-45223-x
A Hybrid Gene Selection Method Based on ReliefF and Ant Colony Optimization Algorithm for Tumor Classification
Abstract
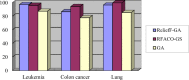
For the DNA microarray datasets, tumor classification based on gene expression profiles has drawn great attention, and gene selection plays a significant role in improving the classification performance of microarray data. In this study, an effective hybrid gene selection method based on ReliefF and Ant colony optimization (ACO) algorithm for tumor classification is proposed. First, for the ReliefF algorithm, the average distance among k nearest or k non-nearest neighbor samples are introduced to estimate the difference among samples, based on which the distances between the samples in the same class or the different classes are defined, and then it can more effectively evaluate the weight values of genes for samples. To obtain the stable results in emergencies, a distance coefficient is developed to construct a new formula of updating weight coefficient of genes to further reduce the instability during calculations. When decreasing the distance between the same samples and increasing the distance between the different samples, the weight division is more obvious. Thus, the ReliefF algorithm can be improved to reduce the initial dimensionality of gene expression datasets and obtain a candidate gene subset. Second, a new pruning rule is designed to reduce dimensionality and obtain a new candidate subset with the smaller number of genes. The probability formula of the next point in the path selected by the ants is presented to highlight the closeness of the correlation relationship between the reaction variables. To increase the pheromone concentration of important genes, a new phenotype updating formula of the ACO algorithm is adopted to prevent the pheromone left by the ants that are overwhelmed with time, and then the weight coefficients of the genes are applied here to eliminate the interference of difference data as much as possible. It follows that the improved ACO algorithm has the ability of the strong positive feedback, which quickly converges to an optimal solution through the accumulation and the updating of pheromone. Finally, by combining the improved ReliefF algorithm and the improved ACO method, a hybrid filter-wrapper-based gene selection algorithm called as RFACO-GS is proposed. The experimental results under several public gene expression datasets demonstrate that the proposed method is very effective, which can significantly reduce the dimensionality of gene expression datasets, and select the most relevant genes with high classification accuracy.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
A hybrid gene selection algorithm based on interaction information for microarray-based cancer classification.PLoS One. 2019 Feb 15;14(2):e0212333. doi: 10.1371/journal.pone.0212333. eCollection 2019. PLoS One. 2019. PMID: 30768654 Free PMC article.
-
A modified ant colony optimization algorithm for tumor marker gene selection.Genomics Proteomics Bioinformatics. 2009 Dec;7(4):200-8. doi: 10.1016/S1672-0229(08)60050-9. Genomics Proteomics Bioinformatics. 2009. PMID: 20172493 Free PMC article.
-
A hybrid gene selection approach for microarray data classification using cellular learning automata and ant colony optimization.Genomics. 2016 Jun;107(6):231-8. doi: 10.1016/j.ygeno.2016.05.001. Epub 2016 May 3. Genomics. 2016. PMID: 27154739
-
Filter versus wrapper gene selection approaches in DNA microarray domains.Artif Intell Med. 2004 Jun;31(2):91-103. doi: 10.1016/j.artmed.2004.01.007. Artif Intell Med. 2004. PMID: 15219288 Review.
-
Optimal features selection in the high dimensional data based on robust technique: Application to different health database.Heliyon. 2024 Sep 2;10(17):e37241. doi: 10.1016/j.heliyon.2024.e37241. eCollection 2024 Sep 15. Heliyon. 2024. PMID: 39296019 Free PMC article. Review.
Cited by
-
Statistical Approach for Biologically Relevant Gene Selection from High-Throughput Gene Expression Data.Entropy (Basel). 2020 Oct 25;22(11):1205. doi: 10.3390/e22111205. Entropy (Basel). 2020. PMID: 33286973 Free PMC article.
-
Application of Swarm Intelligence Optimization Algorithms in Image Processing: A Comprehensive Review of Analysis, Synthesis, and Optimization.Biomimetics (Basel). 2023 Jun 3;8(2):235. doi: 10.3390/biomimetics8020235. Biomimetics (Basel). 2023. PMID: 37366829 Free PMC article. Review.
-
A graph-based gene selection method for medical diagnosis problems using a many-objective PSO algorithm.BMC Med Inform Decis Mak. 2021 Nov 27;21(1):333. doi: 10.1186/s12911-021-01696-3. BMC Med Inform Decis Mak. 2021. PMID: 34838034 Free PMC article.
-
A voting-based machine learning approach for classifying biological and clinical datasets.BMC Bioinformatics. 2023 Apr 11;24(1):140. doi: 10.1186/s12859-023-05274-4. BMC Bioinformatics. 2023. PMID: 37041456 Free PMC article.
-
Machine Learning Based Computational Gene Selection Models: A Survey, Performance Evaluation, Open Issues, and Future Research Directions.Front Genet. 2020 Dec 10;11:603808. doi: 10.3389/fgene.2020.603808. eCollection 2020. Front Genet. 2020. PMID: 33362861 Free PMC article. Review.
References
-
- Greenman CD. Haploinsufficient gene selection in cancer. Science. 2012;337(6090):47–48. - PubMed
-
- Sun L, et al. Joint neighborhood entropy-based gene selection method with fisher score for tumor classification. Applied Intelligence. 2019;49(4):1245–1259.
-
- Cao J, Zhang L, Wang BJ, Li F, Yang J. A fast gene selection method for multi-cancer classification using multiple support vector data description. Journal of Biomedical Informatics. 2015;53:381–389. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

