Computational analysis of single-cell transcriptomics data elucidates the stabilization of Oct4 expression in the E3.25 mouse preimplantation embryo
- PMID: 31222057
- PMCID: PMC6586892
- DOI: 10.1038/s41598-019-45438-y
Computational analysis of single-cell transcriptomics data elucidates the stabilization of Oct4 expression in the E3.25 mouse preimplantation embryo
Abstract
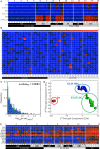
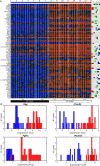
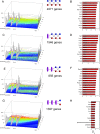
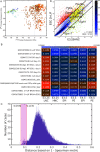
Our computational analysis focuses on the 32- to 64-cell mouse embryo transition, Embryonic day (E3.25), whose study in literature is concentrated mainly on the search for an early onset of the second cell-fate decision, the specification of the inner cell mass (ICM) to primitive endoderm (PE) and epiblast (EPI). We analysed single-cell (sc) microarray transcriptomics data from E3.25 using Hierarchical Optimal k-Means (HOkM) clustering, and identified two groups of ICM cells: a group of cells from embryos with less than 34 cells (E3.25-LNCs), and another group of cells from embryos with more than 33 cells (E3.25-HNCs), corresponding to two developmental stages. Although we found massive underlying heterogeneity in the ICM cells at E3.25-HNC with over 3,800 genes with transcriptomics bifurcation, many of which are PE and EPI markers, we showed that the E3.25-HNCs are neither PE nor EPI. Importantly, analysing the differently expressed genes between the E3.25-LNCs and E3.25-HNCs, we uncovered a non-autonomous mechanism, based on a minimal number of four inner-cell contacts in the ICM, which activates Oct4 in the preimplantation embryo. Oct4 is highly expressed but unstable at E3.25-LNC, and stabilizes at high level at E3.25-HNC, with Bsg highly expressed, and the chromatin remodelling program initialised to establish an early naïve pluripotent state. Our results indicate that the pluripotent state we found to exist in the ICM at E3.25-HNC is the in vivo counterpart of a new, very early pluripotent state. We compared the transcriptomics profile of this in vivo E3.25-HNC pluripotent state, together with the profiles of E3.25-LNC, E3.5 EPI and E4.5 EPI cells, with the profiles of all embryonic stem cells (ESCs) available in the GEO database from the same platform (over 600 microarrays). The shortest distance between the set of inner cells (E3.25, E3.5 and E4.5) and the ESCs is between the E3.25-HNC cells and 2i + LIF ESCs; thus, the developmental transition from 33 to 34 cells decreases dramatically the distance with the naïve ground state of the 2i + LIF ESCs. We validated the E3.25 events through analysis of scRNA-seq data from early and late 32-cell ICM cells.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Does mouse embryo primordial germ cell activation start before implantation as suggested by single-cell transcriptomics dynamics?Mol Hum Reprod. 2016 Mar;22(3):208-25. doi: 10.1093/molehr/gav072. Epub 2016 Jan 5. Mol Hum Reprod. 2016. PMID: 26740066
-
Lineage segregation in human pre-implantation embryos is specified by YAP1 and TEAD1.Hum Reprod. 2023 Aug 1;38(8):1484-1498. doi: 10.1093/humrep/dead107. Hum Reprod. 2023. PMID: 37295962
-
HIPPO pathway members restrict SOX2 to the inner cell mass where it promotes ICM fates in the mouse blastocyst.PLoS Genet. 2014 Oct 23;10(10):e1004618. doi: 10.1371/journal.pgen.1004618. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25340657 Free PMC article.
-
Transcription regulation and chromatin structure in the pluripotent ground state.Biochim Biophys Acta. 2014 Mar;1839(3):129-37. doi: 10.1016/j.bbagrm.2013.09.005. Epub 2013 Oct 2. Biochim Biophys Acta. 2014. PMID: 24096207 Review.
-
Uncovering the true identity of naïve pluripotent stem cells.Trends Cell Biol. 2013 Sep;23(9):442-8. doi: 10.1016/j.tcb.2013.04.004. Epub 2013 May 17. Trends Cell Biol. 2013. PMID: 23685019 Review.
Cited by
-
An Integrative Omics Approach Reveals Involvement of BRCA1 in Hepatic Metastatic Progression of Colorectal Cancer.Cancers (Basel). 2020 Aug 22;12(9):2380. doi: 10.3390/cancers12092380. Cancers (Basel). 2020. PMID: 32842712 Free PMC article.
-
Protective Vaccination Reshapes Hepatic Response to Blood-Stage Malaria of Genes Preferentially Expressed by NK Cells.Vaccines (Basel). 2020 Nov 13;8(4):677. doi: 10.3390/vaccines8040677. Vaccines (Basel). 2020. PMID: 33202767 Free PMC article.
-
Of numbers and movement - understanding transcription factor pathogenesis by advanced microscopy.Dis Model Mech. 2020 Dec 29;13(12):dmm046516. doi: 10.1242/dmm.046516. Dis Model Mech. 2020. PMID: 33433399 Free PMC article. Review.
-
FOntCell: Fusion of Ontologies of Cells.Front Cell Dev Biol. 2021 Feb 11;9:562908. doi: 10.3389/fcell.2021.562908. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33644039 Free PMC article.
-
The Wnt/TCF7L1 transcriptional repressor axis drives primitive endoderm formation by antagonizing naive and formative pluripotency.Nat Commun. 2023 Mar 3;14(1):1210. doi: 10.1038/s41467-023-36914-1. Nat Commun. 2023. PMID: 36869101 Free PMC article.
References
-
- Zernicka-Goetz M, Morris SA, Bruce AW. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat. Rev. Genet. 2009;10:467–477. - PubMed
-
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

