Post-Transcriptional Noise Control
- PMID: 31222776
- PMCID: PMC6637019
- DOI: 10.1002/bies.201900044
Post-Transcriptional Noise Control
Abstract
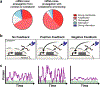
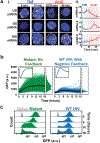
Recent evidence indicates that transcriptional bursts are intrinsically amplified by messenger RNA cytoplasmic processing to generate large stochastic fluctuations in protein levels. These fluctuations can be exploited by cells to enable probabilistic bet-hedging decisions. But large fluctuations in gene expression can also destabilize cell-fate commitment. Thus, it is unclear if cells temporally switch from high to low noise, and what mechanisms enable this switch. Here, the discovery of a post-transcriptional mechanism that attenuates noise in HIV is reviewed. Early in its life cycle, HIV amplifies transcriptional fluctuations to probabilistically select alternate fates, whereas at late times, HIV utilizes a post-transcriptional feedback mechanism to commit to a specific fate. Reanalyzing various reported post-transcriptional negative feedback architectures reveals that they attenuate noise more efficiently than classic transcriptional autorepression, leading to the derivation of an assay to detect post-transcriptional motifs. It is hypothesized that coupling transcriptional and post-transcriptional autoregulation enables efficient temporal noise control to benefit developmental bet-hedging decisions.
Keywords: autoregulation; fate selection; negative feedback; noise control; post-transcriptional; splicing; stochastic noise.
© 2019 WILEY Periodicals, Inc.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

