Enterotype Variations of the Healthy Human Gut Microbiome in Different Geographical Regions
- PMID: 31223215
- PMCID: PMC6563668
- DOI: 10.6026/97320630014560
Enterotype Variations of the Healthy Human Gut Microbiome in Different Geographical Regions
Abstract
Enterotypes are used for classifying individuals based on the gut microbiome. A number of studies are available to find the Enterotypes in healthy individuals; however, most of them lack comparisons at the world level. We analyzed the healthy human gut microbiomes of 495 datasets available in the European Nucleotide Archive (ENA) database derived from fifteen countries from four continents. Firmicutes and Bacteroidetes were the two most abundant phyla in the healthy human gut, worldwide. A high ratio of Proteobacteriato Actinobacteria and a low abundance of Prevotella were identified as the indicators of IBD. Prevotella, Bacteroides, and Bifidobacterium were identified as the Enterotypes in the inter-continental comparisons. At the intra-continental level, two (Bacteroides and Ruminococcaceae), four (Faecalibacterium, Bacteroides, Prevotella, and Clostridiales), and two (Prevotella, Bacteroides/Bifidobacterium) Enterotypes were identified in the American, European, and Asian continents, respectively. In addition, a high abundance of the unknown genus of Ruminococcaeae was observed in the Colombian human gut microbiome. A substantial impact of the geographical distance was observed on human gut microbiome variations, demonstrating a cumulative effect of factors, including dietary habits, genetics, lifestyle, environment, and climate, etc.
Keywords: Enterotype; geographical factor; healthy human gut microbiome; inter-continental.
Figures
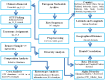
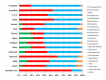
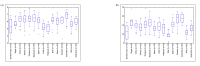



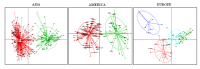
References
LinkOut - more resources
Full Text Sources
