Recent progress in the role of autophagy in neurological diseases
- PMID: 31225510
- PMCID: PMC6551859
- DOI: 10.15698/cst2019.05.186
Recent progress in the role of autophagy in neurological diseases
Abstract
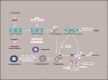
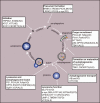
Autophagy (here refers to macroautophagy) is a catabolic pathway by which large protein aggregates and damaged organelles are first sequestered into a double-membraned structure called autophago-some and then delivered to lysosome for destruction. Recently, tremen-dous progress has been made to elucidate the molecular mechanism and functions of this essential cellular metabolic process. In addition to being either a rubbish clearing system or a cellular surviving program in response to different stresses, autophagy plays important roles in a large number of pathophysiological conditions, such as cancer, diabetes, and especially neurodegenerative disorders. Here we review recent progress in the role of autophagy in neurological diseases and discuss how dysregulation of autophagy initiation, autophagosome formation, maturation, and/or au-tophagosome-lysosomal fusion step contributes to the pathogenesis of these disorders in the nervous system.
Keywords: Alzheimer's disease; Amyotrophic lateral sclerosis; Huntington's disease; Parkinson's disease; autophagy; mTOR; neuro-degenerative diseases.
Conflict of interest statement
Conflict of interest: The authors declare that they have no conflict of interests.
Figures
Similar articles
-
Autophagy and apoptosis dysfunction in neurodegenerative disorders.Prog Neurobiol. 2014 Jan;112:24-49. doi: 10.1016/j.pneurobio.2013.10.004. Epub 2013 Nov 6. Prog Neurobiol. 2014. PMID: 24211851 Review.
-
Exploring the Role of Autophagy Dysfunction in Neurodegenerative Disorders.Mol Neurobiol. 2021 Oct;58(10):4886-4905. doi: 10.1007/s12035-021-02472-0. Epub 2021 Jul 2. Mol Neurobiol. 2021. PMID: 34212304 Review.
-
Autophagosome dynamics in neurodegeneration at a glance.J Cell Sci. 2015 Apr 1;128(7):1259-67. doi: 10.1242/jcs.161216. J Cell Sci. 2015. PMID: 25829512 Free PMC article. Review.
-
Recent advances in autophagy-based neuroprotection.Expert Rev Neurother. 2015 Feb;15(2):195-205. doi: 10.1586/14737175.2015.1002087. Expert Rev Neurother. 2015. PMID: 25614954 Review.
-
Role of autophagy in the pathogenesis of amyotrophic lateral sclerosis.Biochim Biophys Acta. 2015 Nov;1852(11):2517-24. doi: 10.1016/j.bbadis.2015.08.005. Epub 2015 Aug 8. Biochim Biophys Acta. 2015. PMID: 26264610 Review.
Cited by
-
Molecular Pathogenicity of Enteroviruses Causing Neurological Disease.Front Microbiol. 2020 Apr 9;11:540. doi: 10.3389/fmicb.2020.00540. eCollection 2020. Front Microbiol. 2020. PMID: 32328043 Free PMC article. Review.
-
mTOR in Alzheimer disease and its earlier stages: Links to oxidative damage in the progression of this dementing disorder.Free Radic Biol Med. 2021 Jun;169:382-396. doi: 10.1016/j.freeradbiomed.2021.04.025. Epub 2021 Apr 30. Free Radic Biol Med. 2021. PMID: 33933601 Free PMC article. Review.
-
Autophagy in health and disease: From molecular mechanisms to therapeutic target.MedComm (2020). 2022 Jul 10;3(3):e150. doi: 10.1002/mco2.150. eCollection 2022 Sep. MedComm (2020). 2022. PMID: 35845350 Free PMC article. Review.
-
In vitro methods in autophagy research: Applications in neurodegenerative diseases and mood disorders.Front Mol Neurosci. 2023 Apr 12;16:1168948. doi: 10.3389/fnmol.2023.1168948. eCollection 2023. Front Mol Neurosci. 2023. PMID: 37122628 Free PMC article. Review.
-
A narrative review of autophagy in migraine.Front Neurosci. 2025 Feb 14;19:1500189. doi: 10.3389/fnins.2025.1500189. eCollection 2025. Front Neurosci. 2025. PMID: 40027467 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
