Cross-transmission Is Not the Source of New Mycobacterium abscessus Infections in a Multicenter Cohort of Cystic Fibrosis Patients
- PMID: 31225586
- PMCID: PMC7156781
- DOI: 10.1093/cid/ciz526
Cross-transmission Is Not the Source of New Mycobacterium abscessus Infections in a Multicenter Cohort of Cystic Fibrosis Patients
Abstract
Background: Mycobacterium abscessus is an extensively drug-resistant pathogen that causes pulmonary disease, particularly in cystic fibrosis (CF) patients. Identifying direct patient-to-patient transmission of M. abscessus is critically important in directing an infection control policy for the management of risk in CF patients. A variety of clinical labs have used molecular epidemiology to investigate transmission. However, there is still conflicting evidence as to how M. abscessus is acquired and whether cross-transmission occurs. Recently, labs have applied whole-genome sequencing (WGS) to investigate this further and, in this study, we investigated whether WGS can reliably identify cross-transmission in M. abscessus.
Methods: We retrospectively sequenced the whole genomes of 145 M. abscessus isolates from 62 patients, seen at 4 hospitals in 2 countries over 16 years.
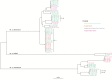
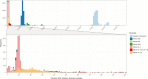
Results: We have shown that a comparison of a fixed number of core single nucleotide variants alone cannot be used to infer cross-transmission in M. abscessus but does provide enough information to replace multiple existing molecular assays. We detected 1 episode of possible direct patient-to-patient transmission in a sibling pair. We found that patients acquired unique M. abscessus strains even after spending considerable time on the same wards with other M. abscessus-positive patients.
Conclusions: This novel analysis has demonstrated that the majority of patients in this study have not acquired M. abscessus through direct patient-to-patient transmission or a common reservoir. Tracking transmission using WGS will only realize its full potential with proper environmental screening, as well as patient sampling.
Keywords: cystic fibrosis; nontuberculous mycobacteria; phylogenomics; transmission; whole-genome sequencing.
© The Author(s) 2019. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures



References
-
- Griffith DE, Brown-Elliott BA, Benwill JL, Wallace RJ Jr. Mycobacterium abscessus. “Pleased to meet you, hope you guess my name.” Ann Am Thorac Soc 2015; 12:436–9. - PubMed
-
- Robinson PD, Harris KA, Aurora P, Hartley JC, Tsang V, Spencer H. Paediatric lung transplant outcomes vary with Mycobacterium abscessus complex species. Eur Respir J 2013; 41:1230–2. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

