For common community phylogenetic analyses, go ahead and use synthesis phylogenies
- PMID: 31225900
- PMCID: PMC7079099
- DOI: 10.1002/ecy.2788
For common community phylogenetic analyses, go ahead and use synthesis phylogenies
Abstract
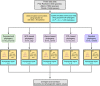
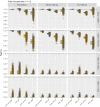
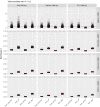
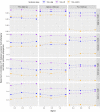
Should we build our own phylogenetic trees based on gene sequence data, or can we simply use available synthesis phylogenies? This is a fundamental question that any study involving a phylogenetic framework must face at the beginning of the project. Building a phylogeny from gene sequence data (purpose-built phylogeny) requires more effort, expertise, and cost than subsetting an already available phylogeny (synthesis-based phylogeny). However, we still lack a comparison of how these two approaches to building phylogenetic trees influence common community phylogenetic analyses such as comparing community phylogenetic diversity and estimating trait phylogenetic signal. Here, we generated three purpose-built phylogenies and their corresponding synthesis-based trees (two from Phylomatic and one from the Open Tree of Life, OTL). We simulated 1,000 communities and 12,000 continuous traits along each purpose-built phylogeny. We then compared the effects of different trees on estimates of phylogenetic diversity (alpha and beta) and phylogenetic signal (Pagel's λ and Blomberg's K). Synthesis-based phylogenies generally yielded higher estimates of phylogenetic diversity when compared to purpose-built phylogenies. However, resulting measures of phylogenetic diversity from both types of phylogenies were highly correlated (Spearman's > 0.8 in most cases). Mean pairwise distance (both alpha and beta) is the index that is most robust to the differences in tree construction that we tested. Measures of phylogenetic diversity based on the OTL showed the highest correlation with measures based on the purpose-built phylogenies. Trait phylogenetic signal estimated with synthesis-based phylogenies, especially from the OTL, was also highly correlated with estimates of Blomberg's K or close to Pagel's λ from purpose-built phylogenies when traits were simulated under Brownian motion. For commonly employed community phylogenetic analyses, our results justify taking advantage of recently developed and continuously improving synthesis trees, especially the Open Tree of Life.
Keywords: alpha diversity; beta diversity; community phylogenetic structure; open tree of life; phylogenetic diversity; phylogenetic signal; trait.
© 2019 by the Ecological Society of America.
Figures


References
-
- Antonelli, A. , Hettling H., Condamine F. L., Vos K., Nilsson R. H., Sanderson M. J., Sauquet H., Scharn R., Silvestro D., and Töpel M.. 2017. Toward a self‐updating platform for estimating rates of speciation and migration, ages, and relationships of taxa. Systematic Biology 66:152–166. - PMC - PubMed
-
- APG III . 2009. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society 161:105–121.
-
- APG IV et al. 2016. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society 181:1–20.
-
- Baum, D. A. , and Smith S. D.. 2012. Tree thinking: An introduction to phylogenetic biology. Roberts Co., Greenwood Village, Colorado, USA.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
