Creating a Synthetic Clinical Trial: Comparative Effectiveness Analyses Using an Electronic Medical Record
- PMID: 31225984
- PMCID: PMC6874028
- DOI: 10.1200/CCI.19.00037
Creating a Synthetic Clinical Trial: Comparative Effectiveness Analyses Using an Electronic Medical Record
Abstract
Purpose: Electronic medical records (EMRs) are a vast resource of potentially mineable data that can be used to complement and extend clinical trials. Extracting and analyzing EMR data are impeded by technical complexities associated with large, multiformat databases. We sought to develop and validate a framework that would overcome the difficulties associated with EMR data and create a simple, portable, and expandable system to better use this resource.
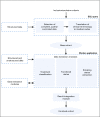
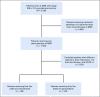
Materials and methods: An Internet-accessible program was developed in Python that applied user-defined criteria to identify and extract patient data from Memorial Sloan Kettering databases. A Worker Application composed of individual modules was developed to identify each patient's functional status, smoking status, and treatment classification. The validity of this approach was tested by identifying, extracting, and analyzing data from a patient cohort that paralleled a practice-changing, prospective, randomized phase III clinical trial performed at a different institution. We called this a synthetic clinical trial.
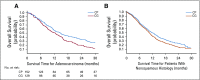
Results: Our synthetic clinical trial identified and extracted data on a cohort of 281 patients with lung cancer who matched inclusion criteria and received their first treatment between October 2003 and July 2010. The data extraction modules were precise and accurate, with F-measures greater than 0.98. Results were similar in directionality and magnitude to the chosen comparator clinical trial.
Conclusion: Our framework offers an accurate and user-friendly interface for identifying and extracting EMR data that can be used to create synthetic clinical trials. Additional studies are needed to validate this approach in other patient cohorts, replicate our findings, and leverage this methodology to improve patient care and accelerate drug development.
Conflict of interest statement
Marjorie G. Zauderer
Isaac Wagner
Aryeh Caroline
Mark G. Kris
No other potential conflicts of interest were reported.
Figures

References
-
- Franklin JM, Schneeweiss S. When and how can real world data analyses substitute for randomized controlled trials? Clin Pharmacol Ther. 2017;102:924–933. - PubMed
-
- Genestreti G, Giovannini N, Frizziero M, et al. Carboplatin and gemcitabine in first-line treatment of elderly patients with advanced non-small cell lung cancer: Data from a retrospective study. J Chemother. 2011;23:232–237. - PubMed
-
- Samelis GF, Ekmektzoglou KA, Tsiakou A, et al. Survival benefit during zoledronic acid and docetaxel-based chemotherapy in metastatic hormone-refractory prostate cancer patients: An institutional report. J BUON. 2011;16:738–743. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

