Nucleotide excision repair of abasic DNA lesions
- PMID: 31226203
- PMCID: PMC6895268
- DOI: 10.1093/nar/gkz558
Nucleotide excision repair of abasic DNA lesions
Abstract
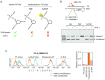
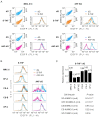
Apurinic/apyrimidinic (AP) sites are a class of highly mutagenic and toxic DNA lesions arising in the genome from a number of exogenous and endogenous sources. Repair of AP lesions takes place predominantly by the base excision pathway (BER). However, among chemically heterogeneous AP lesions formed in DNA, some are resistant to the endonuclease APE1 and thus refractory to BER. Here, we employed two types of reporter constructs accommodating synthetic APE1-resistant AP lesions to investigate the auxiliary repair mechanisms in human cells. By combined analyses of recovery of the transcription rate and suppression of transcriptional mutagenesis at specifically positioned AP lesions, we demonstrate that nucleotide excision repair pathway (NER) efficiently removes BER-resistant AP lesions and significantly enhances the repair of APE1-sensitive ones. Our results further indicate that core NER components XPA and XPF are equally required and that both global genome (GG-NER) and transcription coupled (TC-NER) subpathways contribute to the repair.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993; 362:709–715. - PubMed
-
- Loeb L.A., Preston B.D.. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 1986; 20:201–230. - PubMed
-
- Barnes D.E., Lindahl T.. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004; 38:445–476. - PubMed
-
- Mol C.D., Hosfield D.J., Tainer J.A.. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat. Res. 2000; 460:211–229. - PubMed
-
- Fung H., Demple B.. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell. 2005; 17:463–470. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

