Genetic studies of abdominal MRI data identify genes regulating hepcidin as major determinants of liver iron concentration
- PMID: 31226389
- PMCID: PMC6694204
- DOI: 10.1016/j.jhep.2019.05.032
Genetic studies of abdominal MRI data identify genes regulating hepcidin as major determinants of liver iron concentration
Abstract
Background & aims: Excess liver iron content is common and is linked to the risk of hepatic and extrahepatic diseases. We aimed to identify genetic variants influencing liver iron content and use genetics to understand its link to other traits and diseases.
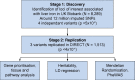
Methods: First, we performed a genome-wide association study (GWAS) in 8,289 individuals from UK Biobank, whose liver iron level had been quantified by magnetic resonance imaging, before validating our findings in an independent cohort (n = 1,513 from IMI DIRECT). Second, we used Mendelian randomisation to test the causal effects of 25 predominantly metabolic traits on liver iron content. Third, we tested phenome-wide associations between liver iron variants and 770 traits and disease outcomes.
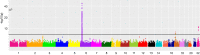
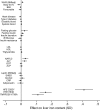
Results: We identified 3 independent genetic variants (rs1800562 [C282Y] and rs1799945 [H63D] in HFE and rs855791 [V736A] in TMPRSS6) associated with liver iron content that reached the GWAS significance threshold (p <5 × 10-8). The 2 HFE variants account for ∼85% of all cases of hereditary haemochromatosis. Mendelian randomisation analysis provided evidence that higher central obesity plays a causal role in increased liver iron content. Phenome-wide association analysis demonstrated shared aetiopathogenic mechanisms for elevated liver iron, high blood pressure, cirrhosis, malignancies, neuropsychiatric and rheumatological conditions, while also highlighting inverse associations with anaemias, lipidaemias and ischaemic heart disease.
Conclusion: Our study provides genetic evidence that mechanisms underlying higher liver iron content are likely systemic rather than organ specific, that higher central obesity is causally associated with higher liver iron, and that liver iron shares common aetiology with multiple metabolic and non-metabolic diseases.
Lay summary: Excess liver iron content is common and is associated with liver diseases and metabolic diseases including diabetes, high blood pressure, and heart disease. We identified 3 genetic variants that are linked to an increased risk of developing higher liver iron content. We show that the same genetic variants are linked to higher risk of many diseases, but they may also be associated with some health advantages. Finally, we use genetic variants associated with waist-to-hip ratio as a tool to show that central obesity is causally associated with increased liver iron content.
Keywords: Genetics; Genome-wide association study; Iron; Magnetic resonance imaging; Metabolic syndrome; Metabolism.
Copyright © 2019 European Association for the Study of the Liver. All rights reserved.
Figures


References
-
- Williams R., Aspinall R., Bellis M., Camps-Walsh G., Cramp M., Dhawan A. The Lancet Commissions Addressing liver disease in the UK: a blueprint for attaining excellence in health care and reducing premature mortality from lifestyle issues of excess consumption of alcohol, obesity, and viral hepatitis. Lancet. 2014;384(9958):1953–1997. - PubMed
-
- Rinella M.E. Nonalcoholic fatty liver disease: a systematic review. JAMA. 2015;313:2263–2273. - PubMed
-
- Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. - PubMed
-
- Pais R., Charlotte F., Fedchuk L., Bedossa P., Lebray P., Poynard T. A systematic review of follow-up biopsies reveals disease progression in patients with non-alcoholic fatty liver. J Hepatol. 2013;59:550–556. - PubMed
-
- Valenti L., Fracanzani A.L., Bugianesi E., Dongiovanni P., Galmozzi E., Vanni E. HFE genotype, parenchymal iron accumulation, and liver fibrosis in patients with nonalcoholic fatty liver disease. Gastroenterology. 2010;138:905–912. - PubMed

