Proteomic analysis by iTRAQ-PRM provides integrated insight into mechanisms of resistance in pepper to Bemisia tabaci (Gennadius)
- PMID: 31226939
- PMCID: PMC6588876
- DOI: 10.1186/s12870-019-1849-0
Proteomic analysis by iTRAQ-PRM provides integrated insight into mechanisms of resistance in pepper to Bemisia tabaci (Gennadius)
Abstract
Background: The Bemisia tabaci is a major leaf feeding insect pest to pepper (Capsicum annuum), causing serious damage to pepper growth and yield. It is particularly important to study the mechanism of pepper resistance to B. tabaci, and to breed and promote the varieties of pepper resistant to B. tabaci. However, very limited molecular mechanism is available about how plants perceive and defend themselves from the destructive pest. Proteome technologies have provided an idea method for studying plant physiological processes in response to B. tabaci.
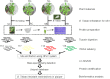
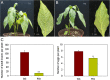
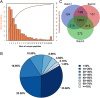
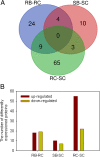
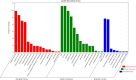
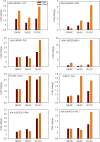
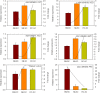
Results: Here, a highly resistant genotype and a highly susceptible genotype were exposed to B. tabaci feeding for 48 h to explore the defense mechanisms of pepper resistance to B. tabaci. The proteomic differences between both genotypes were compared using isobaric tag for relative and absolute quantification (iTRAQ). The quantitative data were validated by parallel reaction monitoring (PRM). The results showed that 37 differential abundance proteins (DAPs) were identified in the RG (resistant genotype), while 17 DAPs were identified in the SG (susceptible genotype) at 48 h after B. tabaci feeding. 77 DAPs were identified when comparing RG with SG without feeding. The DAP functions were determined for the classification of the pathways, mainly involved in redox regulation, stress response, protein metabolism, lipid metabolism and carbon metabolism. Some candidate DAPs are closely related to B. tabaci resistance such as annexin D4-like (ANN4), calreticulin-3 (CRT3), heme-binding protein 2-like (HBP1), acidic endochitinase pcht28-like (PR3) and lipoxygenase 2 (LOX2).
Conclusions: Taken together, this study indicates complex resistance-related events in B. tabaci interaction, provides novel insights into the molecular mechanism underlying the response of plant to B. tabaci, and identifies some candidate proteins against B. tabaci attack.
Keywords: Bemisia tabaci; PRM; Pepper; Proteome; Resistance; iTRAQ.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Response of Bemisia tabaci Genn. (Hemiptera: Aleyrodidae) biotype B to genotypes of pepper Capsicum annuum (Solanales: Solanaceae).Neotrop Entomol. 2013 Apr;42(2):205-10. doi: 10.1007/s13744-012-0106-0. Epub 2013 Jan 17. Neotrop Entomol. 2013. PMID: 23949756
-
Proteomic analysis of differential anther development from sterile/fertile lines in Capsicum annuum L.PeerJ. 2022 May 27;10:e13168. doi: 10.7717/peerj.13168. eCollection 2022. PeerJ. 2022. PMID: 35651745 Free PMC article.
-
[Effects of host plants on selection behavior and biological parameters of Bemisia tabaci Gennadius biotype B].Ying Yong Sheng Tai Xue Bao. 2009 Sep;20(9):2249-54. Ying Yong Sheng Tai Xue Bao. 2009. PMID: 20030150 Chinese.
-
Influence of plant combinations on population characteristics of Bemisia tabaci biotypes B and Q.J Econ Entomol. 2012 Jun;105(3):930-5. doi: 10.1603/ec10373. J Econ Entomol. 2012. PMID: 22812132
-
Insecticide resistance in Bemisia tabaci Gennadius (Homoptera: Aleyrodidae) and Anopheles gambiae Giles (Diptera: Culicidae) could compromise the sustainability of malaria vector control strategies in West Africa.Acta Trop. 2013 Oct;128(1):7-17. doi: 10.1016/j.actatropica.2013.06.004. Epub 2013 Jun 17. Acta Trop. 2013. PMID: 23792227 Review.
Cited by
-
Regulation of heat shock proteins 70 and their role in plant immunity.J Exp Bot. 2022 Apr 5;73(7):1894-1909. doi: 10.1093/jxb/erab549. J Exp Bot. 2022. PMID: 35022724 Free PMC article. Review.
-
Integrative Omics Analysis of Three Oil Palm Varieties Reveals (Tanzania × Ekona) TE as a Cold-Resistant Variety in Response to Low-Temperature Stress.Int J Mol Sci. 2022 Nov 29;23(23):14926. doi: 10.3390/ijms232314926. Int J Mol Sci. 2022. PMID: 36499255 Free PMC article.
-
Comparative proteomic analysis of silica-induced pulmonary fibrosis in rats based on tandem mass tag (TMT) quantitation technology.PLoS One. 2020 Oct 29;15(10):e0241310. doi: 10.1371/journal.pone.0241310. eCollection 2020. PLoS One. 2020. PMID: 33119648 Free PMC article.
-
Beyond the surface: the plant secretome as a bridge between the cell and its environment.Planta. 2025 Feb 26;261(4):67. doi: 10.1007/s00425-025-04648-7. Planta. 2025. PMID: 40000454 Review.
-
Analysis of Chronic Mild Stress-Induced Hypothalamic Proteome: Identification of Protein Dysregulations Associated With Vulnerability and Resiliency to Depression or Anxiety.Front Mol Neurosci. 2021 Mar 2;14:633398. doi: 10.3389/fnmol.2021.633398. eCollection 2021. Front Mol Neurosci. 2021. PMID: 33737865 Free PMC article.
References
-
- Dinsdale A, Cook L, Riginos C, Buckley YM, Barro PD. Refined global analysis of Bemisia tabaci (hemiptera: sternorrhyncha: aleyrodoidea) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann Entomol Soc Am. 2010;103:196–208. doi: 10.1603/AN09061. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

