Exome-wide assessment of the functional impact and pathogenicity of multinucleotide mutations
- PMID: 31227601
- PMCID: PMC6633265
- DOI: 10.1101/gr.239756.118
Exome-wide assessment of the functional impact and pathogenicity of multinucleotide mutations
Abstract
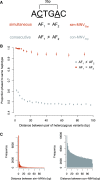
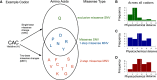
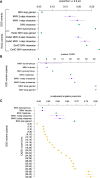
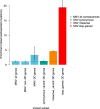
Approximately 2% of de novo single-nucleotide variants (SNVs) appear as part of clustered mutations that create multinucleotide variants (MNVs). MNVs are an important source of genomic variability as they are more likely to alter an encoded protein than a SNV, which has important implications in disease as well as evolution. Previous studies of MNVs have focused on their mutational origins and have not systematically evaluated their functional impact and contribution to disease. We identified 69,940 MNVs and 91 de novo MNVs in 6688 exome-sequenced parent-offspring trios from the Deciphering Developmental Disorders Study comprising families with severe developmental disorders. We replicated the previously described MNV mutational signatures associated with DNA polymerase zeta, an error-prone translesion polymerase, and the APOBEC family of DNA deaminases. We estimate the simultaneous MNV germline mutation rate to be 1.78 × 10-10 mutations per base pair per generation. We found that most MNVs within a single codon create a missense change that could not have been created by a SNV. MNV-induced missense changes were, on average, more physicochemically divergent, were more depleted in highly constrained genes (pLI ≥ 0.9), and were under stronger purifying selection compared with SNV-induced missense changes. We found that de novo MNVs were significantly enriched in genes previously associated with developmental disorders in affected children. This shows that MNVs can be more damaging than SNVs even when both induce missense changes, and are an important variant type to consider in relation to human disease.
© 2019 Kaplanis et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
