IL-33-mediated mast cell activation promotes gastric cancer through macrophage mobilization
- PMID: 31227713
- PMCID: PMC6588585
- DOI: 10.1038/s41467-019-10676-1
IL-33-mediated mast cell activation promotes gastric cancer through macrophage mobilization
Abstract
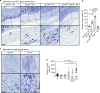
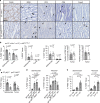
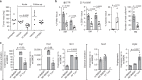
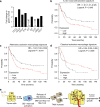
The contribution of mast cells in the microenvironment of solid malignancies remains controversial. Here we functionally assess the impact of tumor-adjacent, submucosal mast cell accumulation in murine and human intestinal-type gastric cancer. We find that genetic ablation or therapeutic inactivation of mast cells suppresses accumulation of tumor-associated macrophages, reduces tumor cell proliferation and angiogenesis, and diminishes tumor burden. Mast cells are activated by interleukin (IL)-33, an alarmin produced by the tumor epithelium in response to the inflammatory cytokine IL-11, which is required for the growth of gastric cancers in mice. Accordingly, ablation of the cognate IL-33 receptor St2 limits tumor growth, and reduces mast cell-dependent production and release of the macrophage-attracting factors Csf2, Ccl3, and Il6. Conversely, genetic or therapeutic macrophage depletion reduces tumor burden without affecting mast cell abundance. Therefore, tumor-derived IL-33 sustains a mast cell and macrophage-dependent signaling cascade that is amenable for the treatment of gastric cancer.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- 1067244/Department of Health | National Health and Medical Research Council (NHMRC)/International
- MR/R026424/1/MRC_/Medical Research Council/United Kingdom
- 1160708/Cancer Council Victoria/International
- 1069024/Department of Health | National Health and Medical Research Council (NHMRC)/International
- 1092788/Department of Health | National Health and Medical Research Council (NHMRC)/International
LinkOut - more resources
Full Text Sources
Medical

