Molecular phenotyping of the surfaceome of migratory chondroprogenitors and mesenchymal stem cells using biotinylation, glycocapture and quantitative LC-MS/MS proteomic analysis
- PMID: 31227739
- PMCID: PMC6588563
- DOI: 10.1038/s41598-019-44957-y
Molecular phenotyping of the surfaceome of migratory chondroprogenitors and mesenchymal stem cells using biotinylation, glycocapture and quantitative LC-MS/MS proteomic analysis
Abstract
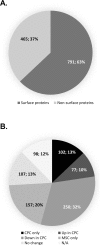
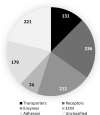
The complement of cell surface proteins, collectively referred to as the surfaceome, is a useful indicator of normal differentiation processes, and the development of pathologies such as osteoarthritis (OA). We employed biochemical and proteomic tools to explore the surfaceome and to define biomarkers in chondrogenic progenitor cells (CPC) derived from human OA knee articular cartilage. These cells have great therapeutic potential, but their unexplored biology limits their clinical application. We performed biotinylation combined with glycocapture and high throughput shotgun proteomics to define the surface proteome of human bone marrow mesenchymal stem cells (MSCs) and human CPCs. We prepared cell surface protein-enriched fractions from MSCs and CPCs, and then a proteomic approach was used to compare and evaluate protein changes between undifferentiated MSCs and CPCs. 1256 proteins were identified in the study, of which 791 (63%) were plasma membrane, cell surface or extracellular matrix proteins. Proteins constituting the surfaceome were annotated and categorized. Our results provide, for the first time, a repository of quantitative proteomic data on the surfaceome of two closely related cell types relevant to cartilage biology and OA. These results may provide novel insights into the transformation of the surfaceome during chondrogenic differentiation and phenotypic changes during OA development.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Josic D, Clifton JG, Kovac S, Hixson DC. Membrane proteins as diagnostic biomarkers and targets for new therapies. Current opinion in molecular therapeutics. 2008;10:116–123. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

