Open chromatin dynamics in prosensory cells of the embryonic mouse cochlea
- PMID: 31227770
- PMCID: PMC6588700
- DOI: 10.1038/s41598-019-45515-2
Open chromatin dynamics in prosensory cells of the embryonic mouse cochlea
Abstract
Hearing loss is often due to the absence or the degeneration of hair cells in the cochlea. Understanding the mechanisms regulating the generation of hair cells may therefore lead to better treatments for hearing disorders. To elucidate the transcriptional control mechanisms specifying the progenitor cells (i.e. prosensory cells) that generate the hair cells and support cells critical for hearing function, we compared chromatin accessibility using ATAC-seq in sorted prosensory cells (Sox2-EGFP+) and surrounding cells (Sox2-EGFP-) from E12, E14.5 and E16 cochlear ducts. In Sox2-EGFP+, we find greater accessibility in and near genes restricted in expression to the prosensory region of the cochlear duct including Sox2, Isl1, Eya1 and Pou4f3. Furthermore, we find significant enrichment for the consensus binding sites of Sox2, Six1 and Gata3-transcription factors required for prosensory development-in the open chromatin regions. Over 2,200 regions displayed differential accessibility with developmental time in Sox2-EGFP+ cells, with most changes in the E12-14.5 window. Open chromatin regions detected in Sox2-EGFP+ cells map to over 48,000 orthologous regions in the human genome that include regions in genes linked to deafness. Our results reveal a dynamic landscape of open chromatin in prosensory cells with potential implications for cochlear development and disease.
Conflict of interest statement
The authors declare no competing interests.
Figures
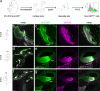


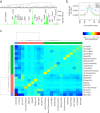

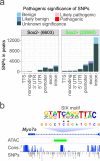
References
Publication types
MeSH terms
Substances
Grants and funding
- R21 DC014768/DC/NIDCD NIH HHS/United States
- T32 GM007270/GM/NIGMS NIH HHS/United States
- T32 DC005361/DC/NIDCD NIH HHS/United States
- T32HD007183/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)/International
- F32 DC016480/DC/NIDCD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

