Noncoding RNA transcription at enhancers and genome folding in cancer
- PMID: 31228211
- PMCID: PMC6676135
- DOI: 10.1111/cas.14107
Noncoding RNA transcription at enhancers and genome folding in cancer
Abstract
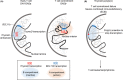
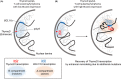
Changes of nuclear localization of lineage-specific genes from a transcriptionally inert to permissive environment are a crucial step in establishing the identity of a cell. Noncoding RNA transcription-mediated genome folding and activation of target gene expression have been found in a variety of cell types. Noncoding RNA ThymoD (thymocyte differentiation factor) transcription at superenhancers is essential for mouse T-cell lineage commitment. The cessation of ThymoD transcription abolishes transcription-mediated demethylation, recruiting looping factors such as the cohesin complex, CCCTC-binding factor (CTCF), ultimately leading to the phenotype of severe combined immunodeficiency and T-cell leukemia/lymphoma. In this review, we describe the functional role of RNA polymerase II-mediated transcription at enhancers and in genome folding. We also highlight the involvement of faulty activation or suppression of enhancer transcription and enhancer-promoter interaction in cancer development.
Keywords: ThymoD; cancer; genome folding; noncoding RNA; transcription.
© 2019 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Figures



References
-
- Kosak ST, Skok JA, Medina KL, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296(5565):158‐162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

