Gene-specific features enhance interpretation of mutational impact on acid α-glucosidase enzyme activity
- PMID: 31228295
- PMCID: PMC7329270
- DOI: 10.1002/humu.23846
Gene-specific features enhance interpretation of mutational impact on acid α-glucosidase enzyme activity
Abstract
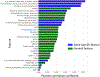
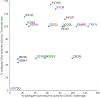
We present a computational model for predicting mutational impact on enzymatic activity of human acid α-glucosidase (GAA), an enzyme associated with Pompe disease. Using a model that combines features specific to GAA with other general evolutionary and physiochemical features, we made blind predictions of enzymatic activity relative to wildtype human GAA for >300 GAA mutants, as part of the Critical Assessment of Genome Interpretation 5 GAA challenge. We found that gene-specific features can improve the performance of existing impact prediction tools that mostly rely on general features for pathogenicity prediction. Majority of the poorly predicted mutants that lower wildtype GAA enzyme activity occurred on the surface of the GAA protein. We also found that gene-specific features were uncorrelated with existing methods and provided orthogonal information for interpreting the origin of pathogenicity, particular in variants that are poorly predicted by existing general methods. Specific variants in GAA, when investigated in the context of its protein structure, suggested gene-specific information like the disruption of local backbone torsional geometry and disruption of particular sidechain-sidechain hydrogen bonds as some potential sources for pathogenicity.
Keywords: Pompe disease; acid α-glucosidase; enzyme activity prediction; gene-specific variant effect prediction; variant interpretation.
© 2019 Wiley Periodicals, Inc.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

