Role of deep learning in infant brain MRI analysis
- PMID: 31229667
- PMCID: PMC6874895
- DOI: 10.1016/j.mri.2019.06.009
Role of deep learning in infant brain MRI analysis
Abstract
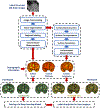
Deep learning algorithms and in particular convolutional networks have shown tremendous success in medical image analysis applications, though relatively few methods have been applied to infant MRI data due numerous inherent challenges such as inhomogenous tissue appearance across the image, considerable image intensity variability across the first year of life, and a low signal to noise setting. This paper presents methods addressing these challenges in two selected applications, specifically infant brain tissue segmentation at the isointense stage and presymptomatic disease prediction in neurodevelopmental disorders. Corresponding methods are reviewed and compared, and open issues are identified, namely low data size restrictions, class imbalance problems, and lack of interpretation of the resulting deep learning solutions. We discuss how existing solutions can be adapted to approach these issues as well as how generative models seem to be a particularly strong contender to address them.
Keywords: Convolutional neural networks; Deep learning; Infant MRI; Isointense segmentation; MRI; Machine learning; Prediction.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures



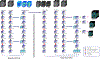






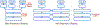

References
-
- A. P. Association, et al., American psychiatric association dsm-5 development, Proposed Revisions/Somatic Symptom Disorders/J 2.
-
- Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM, Fsl, Neuroimage 62 (2) (2012) 782–790. - PubMed
-
- Dai Y, Shi F, Wang L, Wu G, Shen D, ibeat: a toolbox for infant brain magnetic resonance image processing, Neuroinformatics 11 (2) (2013) 211–225. - PubMed
-
- Penny WD, Friston KJ, Ashburner JT, Kiebel SJ, Nichols TE, Statistical parametric mapping: the analysis of functional brain images, Elsevier, 2011.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

