De Novo Variants in TAOK1 Cause Neurodevelopmental Disorders
- PMID: 31230721
- PMCID: PMC6612514
- DOI: 10.1016/j.ajhg.2019.05.005
De Novo Variants in TAOK1 Cause Neurodevelopmental Disorders
Abstract
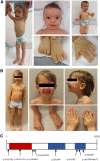
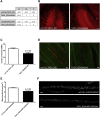
De novo variants represent a significant cause of neurodevelopmental delay and intellectual disability. A genetic basis can be identified in only half of individuals who have neurodevelopmental disorders (NDDs); this indicates that additional causes need to be elucidated. We compared the frequency of de novo variants in patient-parent trios with (n = 2,030) versus without (n = 2,755) NDDs. We identified de novo variants in TAOK1 (thousand and one [TAO] amino acid kinase 1), which encodes the serine/threonine-protein kinase TAO1, in three individuals with NDDs but not in persons who did not have NDDs. Through further screening and the use of GeneMatcher, five additional individuals with NDDs were found to have de novo variants. All eight variants were absent from gnomAD (Genome Aggregation Database). The variant carriers shared a non-specific phenotype of developmental delay, and six individuals had additional muscular hypotonia. We established a fibroblast line of one mutation carrier, and we demonstrated that reduced mRNA levels of TAOK1 could be increased upon cycloheximide treatment. These results indicate nonsense-mediated mRNA decay. Further, there was neither detectable phosphorylated TAO1 kinase nor phosphorylated tau in these cells, and mitochondrial morphology was altered. Knockdown of the ortholog gene Tao1 (Tao, CG14217) in Drosophila resulted in delayed early development. The majority of the Tao1-knockdown flies did not survive beyond the third instar larval stage. When compared to control flies, Tao1 knockdown flies revealed changed morphology of the ventral nerve cord and the neuromuscular junctions as well as a decreased number of endings (boutons). Furthermore, mitochondria in mutant flies showed altered distribution and decreased size in axons of motor neurons. Thus, we provide compelling evidence that de novo variants in TAOK1 cause NDDs.
Keywords: TAO kinase 1; de novo variants; fly model; neurodevelopmental disorders.
Copyright © 2019 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Sheridan E., Wright J., Small N., Corry P.C., Oddie S., Whibley C., Petherick E.S., Malik T., Pawson N., McKinney P.A., Parslow R.C. Risk factors for congenital anomaly in a multiethnic birth cohort: an analysis of the Born in Bradford study. Lancet. 2013;382:1350–1359. - PubMed
- Sheridan, E., Wright, J., Small, N., Corry, P.C., Oddie, S., Whibley, C., Petherick, E.S., Malik, T., Pawson, N., McKinney, P.A., and Parslow, R.C. (2013). Risk factors for congenital anomaly in a multiethnic birth cohort: an analysis of the Born in Bradford study. Lancet 382, 1350-1359. - PubMed
-
- Hoischen A., Krumm N., Eichler E.E. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat. Neurosci. 2014;17:764–772. - PMC - PubMed
- Hoischen, A., Krumm, N., and Eichler, E.E. (2014). Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat. Neurosci. 17, 764-772. - PMC - PubMed
-
- Ku C.S., Polychronakos C., Tan E.K., Naidoo N., Pawitan Y., Roukos D.H., Mort M., Cooper D.N. A new paradigm emerges from the study of de novo mutations in the context of neurodevelopmental disease. Mol. Psychiatry. 2013;18:141–153. - PubMed
- Ku, C.S., Polychronakos, C., Tan, E.K., Naidoo, N., Pawitan, Y., Roukos, D.H., Mort, M., and Cooper, D.N. (2013). A new paradigm emerges from the study of de novo mutations in the context of neurodevelopmental disease. Mol. Psychiatry 18, 141-153. - PubMed
-
- Wright C.F., McRae J.F., Clayton S., Gallone G., Aitken S., FitzGerald T.W., Jones P., Prigmore E., Rajan D., Lord J., DDD Study Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders. Genet. Med. 2018;20:1216–1223. - PMC - PubMed
- Wright, C.F., McRae, J.F., Clayton, S., Gallone, G., Aitken, S., FitzGerald, T.W., Jones, P., Prigmore, E., Rajan, D., Lord, J., et al.; DDD Study (2018). Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders. Genet. Med. 20, 1216-1223. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

