Bioinformatics-Based Identification of Expanded Repeats: A Non-reference Intronic Pentamer Expansion in RFC1 Causes CANVAS
- PMID: 31230722
- PMCID: PMC6612533
- DOI: 10.1016/j.ajhg.2019.05.016
Bioinformatics-Based Identification of Expanded Repeats: A Non-reference Intronic Pentamer Expansion in RFC1 Causes CANVAS
Abstract
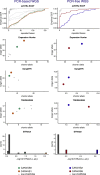
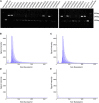
Genomic technologies such as next-generation sequencing (NGS) are revolutionizing molecular diagnostics and clinical medicine. However, these approaches have proven inefficient at identifying pathogenic repeat expansions. Here, we apply a collection of bioinformatics tools that can be utilized to identify either known or novel expanded repeat sequences in NGS data. We performed genetic studies of a cohort of 35 individuals from 22 families with a clinical diagnosis of cerebellar ataxia with neuropathy and bilateral vestibular areflexia syndrome (CANVAS). Analysis of whole-genome sequence (WGS) data with five independent algorithms identified a recessively inherited intronic repeat expansion [(AAGGG)exp] in the gene encoding Replication Factor C1 (RFC1). This motif, not reported in the reference sequence, localized to an Alu element and replaced the reference (AAAAG)11 short tandem repeat. Genetic analyses confirmed the pathogenic expansion in 18 of 22 CANVAS-affected families and identified a core ancestral haplotype, estimated to have arisen in Europe more than twenty-five thousand years ago. WGS of the four RFC1-negative CANVAS-affected families identified plausible variants in three, with genomic re-diagnosis of SCA3, spastic ataxia of the Charlevoix-Saguenay type, and SCA45. This study identified the genetic basis of CANVAS and demonstrated that these improved bioinformatics tools increase the diagnostic utility of WGS to determine the genetic basis of a heterogeneous group of clinically overlapping neurogenetic disorders.
Keywords: CANVAS; ataxia; repeat expansions; short tandem repeats; whole-genome sequencing.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Gymrek M., Willems T., Guilmatre A., Zeng H., Markus B., Georgiev S., Daly M.J., Price A.L., Pritchard J.K., Sharp A.J., Erlich Y. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat. Genet. 2016;48:22–29. - PMC - PubMed
- Gymrek, M., Willems, T., Guilmatre, A., Zeng, H., Markus, B., Georgiev, S., Daly, M.J., Price, A.L., Pritchard, J.K., Sharp, A.J., and Erlich, Y. (2016). Abundant contribution of short tandem repeats to gene expression variation in humans. Nat. Genet. 48, 22-29. - PMC - PubMed
-
- Quilez J., Guilmatre A., Garg P., Highnam G., Gymrek M., Erlich Y., Joshi R.S., Mittelman D., Sharp A.J. Polymorphic tandem repeats within gene promoters act as modifiers of gene expression and DNA methylation in humans. Nucleic Acids Res. 2016;44:3750–3762. - PMC - PubMed
- Quilez, J., Guilmatre, A., Garg, P., Highnam, G., Gymrek, M., Erlich, Y., Joshi, R.S., Mittelman, D., and Sharp, A.J. (2016). Polymorphic tandem repeats within gene promoters act as modifiers of gene expression and DNA methylation in humans. Nucleic Acids Res. 44, 3750-3762. - PMC - PubMed
-
- Subramanian S., Madgula V.M., George R., Mishra R.K., Pandit M.W., Kumar C.S., Singh L. Triplet repeats in human genome: distribution and their association with genes and other genomic regions. Bioinformatics. 2003;19:549–552. - PubMed
- Subramanian, S., Madgula, V.M., George, R., Mishra, R.K., Pandit, M.W., Kumar, C.S., and Singh, L. (2003). Triplet repeats in human genome: distribution and their association with genes and other genomic regions. Bioinformatics 19, 549-552. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

