Cell of Origin and Genetic Alterations in the Pathogenesis of Multiple Myeloma
- PMID: 31231360
- PMCID: PMC6558388
- DOI: 10.3389/fimmu.2019.01121
Cell of Origin and Genetic Alterations in the Pathogenesis of Multiple Myeloma
Abstract
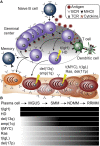
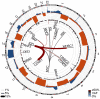
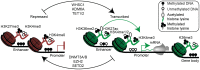
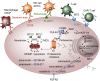
B cell activation and differentiation yields plasma cells with high affinity antibodies to a given antigen in a time-frame that allows for host protection. Although the end product is most commonly humoral immunity, the rapid proliferation and somatic mutation of the B cell receptor also results in oncogenic mutations that cause B cell malignancies including plasma cell neoplasms such as multiple myeloma. Myeloma is the second most common hematological malignancy and results in over 100,000 deaths per year worldwide. The genetic alterations that occur in the germinal center, however, are not sufficient to cause myeloma, but rather impart cell proliferation potential on plasma cells, which are normally non-dividing. This pre-malignant state, referred to as monoclonal gammopathy of undetermined significance or MGUS, provides the opportunity for further genetic and epigenetic alterations eventually resulting in a progressive disease that becomes symptomatic. In this review, we will provide a brief history of clonal gammopathies and detail how some of the key discoveries were interwoven with the study of plasma cells. We will also review the genetic and epigenetic alterations discovered over the past 25 years, how these are instrumental to myeloma pathogenesis, and what these events teach us about myeloma and plasma cell biology. These data will be placed in the context of normal B cell development and differentiation and we will discuss how understanding the biology of plasma cells can lead to more effective therapies targeting multiple myeloma.
Keywords: B cell; IgH translocations; MGUS; MYC; epigenetics; genetics; multiple myeloma; plasma cell.
Figures
References
-
- Jones BH. Chemical pathology. Lancet. (1847) 2:88.
-
- Jones HB. On a New Substance Occurring in the Urine of a Patient with Mollities Ossium. In: Jones HB. editor. Philosophical Transactions of the Royal Society of London. London: Royal Society Stable; (1848) 138:55–62.
-
- Jenner EI. An inquiry into the causes and effects of the variolae vaccinae, or cow-pox. 1798. In: Edward J. editor. The Three Original Publications on Vaccination Against Smallpox. The Harvard Classics (1798). Available online at: http://www.bartleby.com/38/4/1.html (accessed March 22, 2016).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

