CD49a Expression Identifies a Subset of Intrahepatic Macrophages in Humans
- PMID: 31231382
- PMCID: PMC6568245
- DOI: 10.3389/fimmu.2019.01247
CD49a Expression Identifies a Subset of Intrahepatic Macrophages in Humans
Abstract
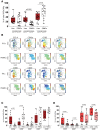
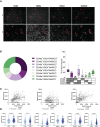
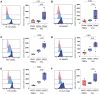
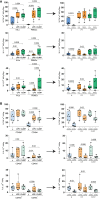
Macrophages play central roles in inflammatory reactions and initiation of immune responses during infections. More than 80% of total tissue macrophages are described to be located in the liver as liver-resident macrophages, also named Kupffer cells (KCs). While studies in mice have established a central role of liver-resident KCs in regulating liver inflammation, their phenotype and function are not well-characterized in humans. Comparing paired human liver and peripheral blood samples, we observed significant differences in the distribution of macrophage (Mφ) subsets, with lower frequencies of CD14hiCD16lo and higher frequencies of CD14int-hiCD16int Mφ in human livers. Intrahepatic Mφ consisted of diverse subsets with differential expression of CD49a, a liver-residency marker previously described for human and mice NK cells, and VSIG4 and/or MARCO, two recently described human tissue Mφ markers. Furthermore, intrahepatic CD49a+ Mφ expressed significantly higher levels of maturation and activation markers, exhibited higher baseline levels of TNF-α, IL-12, and IL-10 production, but responded less to additional in vitro TLR stimulation. In contrast, intrahepatic CD49a- Mφ were highly responsive to stimulation with TLR ligands, similar to what was observed for CD49a- monocytes (MOs) in peripheral blood. Taken together, these studies identified populations of CD49a+, VSIG4+, and/or MARCO+ Mφ in human livers, and demonstrated that intrahepatic CD49a+ Mφ differed in phenotype and function from intrahepatic CD49a- Mφ as well as from peripheral blood-derived monocytes.
Keywords: CD49a; cell activation; human liver; monocytes/macrophages; tissue residency.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

