Enhanced Bone Marrow Homing of Natural Killer Cells Following mRNA Transfection With Gain-of-Function Variant CXCR4R334X
- PMID: 31231387
- PMCID: PMC6560173
- DOI: 10.3389/fimmu.2019.01262
Enhanced Bone Marrow Homing of Natural Killer Cells Following mRNA Transfection With Gain-of-Function Variant CXCR4R334X
Abstract
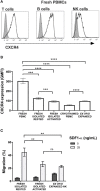
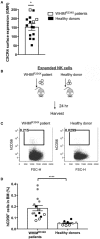
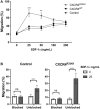
Adoptive transfer of natural killer (NK) cells can induce remission in patients with relapsed/refractory leukemia and myeloma. However, to date, clinical efficacy of NK cell immunotherapy has been limited to a sub-fraction of patients. Here we show that steps incorporated in the ex vivo manipulation/production of NK cell products used for adoptive infusion, such as over-night IL-2 activation or cryopreservation followed by ex vivo expansion, drastically decreases NK cell surface expression of the bone marrow (BM) homing chemokine receptor CXCR4. Reduced CXCR4 expression was associated with dampened in vitro NK cell migration toward its cognate ligand stromal-derived factor-1α (SDF-1α). NK cells isolated from patients with WHIM syndrome carry gain-of-function (GOF) mutations in CXCR4 (CXCR4R334X). Compared to healthy donors, we observed that NK cells expanded from WHIM patients have similar surface levels of CXCR4 but have a much stronger propensity to home to BM compartments when adoptively infused into NOD-scid IL2Rgammanull (NSG) mice. Therefore, in order to augment the capacity of adoptively infused NK cells to home to the BM, we genetically engineered ex vivo expanded NK cells to express the naturally occurring GOF CXCR4R334X receptor variant. Transfection of CXCR4R334X-coding mRNA into ex vivo expanded NK cells using a clinically applicable method consistently led to an increase in cell surface CXCR4 without altering NK cell phenotype, cytotoxic function, or compromising NK cell viability. Compared to non-transfected and wild type CXCR4-coding mRNA transfected counterparts, CXCR4R334X-engineered NK cells had significantly greater chemotaxis toward SDF-1α in vitro. Importantly, expression of CXCR4R334X on expanded NK cells resulted in significantly greater BM homing following adoptive transfer into NSG mice compared to non-transfected NK cell controls. Collectively, these data suggest up-regulation of cell surface CXCR4R334X on ex vivo expanded NK cells via mRNA transfection represents a novel approach to improve homing and target NK cell-based immunotherapies to BM where hematological malignancies reside.
Keywords: CXCR4; NK cells; WHIM; homing; immunotherapy; mRNA transfection.
Figures




References
-
- Bachanova V, Sarhan D, DeFor TE, Cooley S, Panoskaltsis-Mortari A, Blazar BR, et al. . Haploidentical natural killer cells induce remissions in non-Hodgkin lymphoma patients with low levels of immune-suppressor cells. Cancer Immunol Immunother. (2018) 67:483–94. 10.1007/s00262-017-2100-1 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

